Disorders with similar clinical phenotypes reveal underlying genetic interaction: SATB2 acts as an activator of the UPF3B gene - PubMed (original) (raw)
Case Reports
Disorders with similar clinical phenotypes reveal underlying genetic interaction: SATB2 acts as an activator of the UPF3B gene
Petcharat Leoyklang et al. Hum Genet. 2013 Dec.
Abstract
Two syndromic cognitive impairment disorders have very similar craniofacial dysmorphisms. One is caused by mutations of SATB2, a transcription regulator and the other by heterozygous mutations leading to premature stop codons in UPF3B, encoding a member of the nonsense-mediated mRNA decay complex. Here we demonstrate that the products of these two causative genes function in the same pathway. We show that the SATB2 nonsense mutation in our patient leads to a truncated protein that localizes to the nucleus, forms a dimer with wild-type SATB2 and interferes with its normal activity. This suggests that the SATB2 nonsense mutation has a dominant negative effect. The patient's leukocytes had significantly decreased UPF3B mRNA compared to controls. This effect was replicated both in vitro, where siRNA knockdown of SATB2 in HEK293 cells resulted in decreased UPF3B expression, and in vivo, where embryonic tissue of Satb2 knockout mice showed significantly decreased Upf3b expression. Furthermore, chromatin immunoprecipitation demonstrates that SATB2 binds to the UPF3B promoter, and a luciferase reporter assay confirmed that SATB2 expression significantly activates gene transcription using the UPF3B promoter. These findings indicate that SATB2 activates UPF3B expression through binding to its promoter. This study emphasizes the value of recognizing disorders with similar clinical phenotypes to explore underlying mechanisms of genetic interaction.
Figures
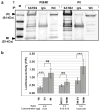
Fig. 1. Protein domains of SATB2 and expression of the wild-type and truncated SATB2 proteins
(a) SATB2 protein outline. SATB2 contains 733 amino acids, including an N-terminal dimerization domain (residues 57–231), two CUT domains with DNA-binding motifs (residues 352–437 and 482–560), a DNA-binding domain (residues 614–677), and a nuclear localization signal (NLS; residues 613–616). The position of our patient’s nonsense mutation, p.R239X, is indicated. (b) Immunoblot analysis. HEK293 cells transfected with the wild-type SATB2/myc_-His plasmid (Wt; 84 kDa), the truncated SATB2/myc_-His plasmid (Mt; 29 kDa), or with the empty pcDNA 3.1/_myc_-His plasmid (EV). Upper panel: Probing with anti c-Myc antibodies (Ab) showed SATB2 bands at the expected molecular weights. Middle panel; Reprobing the membrane with antibodies to the C-terminus of SATB2 showed abundant expression of SATB2 in the Wt SATB2 transfected cells, and less SATB2 expression in the Mt SATB2 and EV transfected cells. Lower panel: Reprobing the same membrane with anti β-actin antibodies demonstrated equal protein loading in all lanes.
Fig. 2. Nuclear localization of the wild-type and truncated SATB2
HEK293 cells were transfected with the Wt (lanes 2–4) or Mt (lanes 5–7) SATB2/_myc_-His plasmids. (a) Immunoblotting of subcellular fractionated cell extracts with anti c-Myc antibodies (Ab) showed that both the Wt and Mt SATB2 proteins were present in the whole cell extracts (WE) and nuclear extracts (NE) and absent from cytoplasmic extracts (CE), upper panel. The membrane was reprobed with antibodies to the C-terminus of SATB2, middle panel and α-tubulin as loading control, lower panel. (b) Immunofluorescence of normal human fibroblasts transfected with Wt or Mt SATB2/_myc_-His plasmids. Cells were stained with rabbit anti c-Myc (red) or mouse anti-SATB2 (green) antibodies, the phalloidin (staining F-actin) to outline cell boundaries (white), and the nuclear dye DAPI (blue). Merged images showed nuclear localization of the Wt and Mt proteins. Note that mock transfected cells, empty vector transfected cells and cells transfected with Mt SATB2/_myc_-His showed endogenous SATB2 upon SATB2 antibody staining.
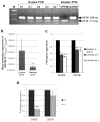
Fig. 3. Interaction between the wild-type and truncated SATB2
(a) HEK293 cells were co-transfected (T) with a combination of the Wt and Mt SATB2/_myc_-His plasmids (Wt&Mt), or with only the Wt SATB2/_myc_-His plasmid (Wt). Cell extracts were immunoprecipitated (IP) with anti-SATB2 or mouse IgG (negative control) antibodies. All IP-fractions and whole cell extracts (WC) were immunoblotted with c-Myc antibodies. The Mt protein was detected in cells transfected with both Wt&Mt SATB2 plasmids (WC fraction) and precipitated with SATB2 antibodies (SATB2 fraction), indicating interaction of the Wt SATB2 with the Mt SATB2. Cells transfected with the Wt plasmid only did not show the 29 kDa protein band, as expected. M = SeeBlue® Plus2 Pre-Stained Standard (Invitrogen). (b) Firefly (F) luciferase activities (from the MAR SATB2 reporter plasmid) shown as fold of Renilla (R) luciferase activity (from normalization plasmid) (F/R, Y-axis) in cells co-transfected with different ratios of the Wt or Mt SATB2/_myc_-His plasmid, or the empty pcDNA3.1/_myc_-His vector (EV). Transfection of the Wt SATB2 repressed MAR-luciferase activity (likely due to repression by SATB2 dimers) compared to EV or Mt transfections. Co-transfections of the Wt with either EV or Mt SATB2 decreased repression (likely due to decreased or aberrant SATB2 dimer formation). The activity of each combination of plasmids was assayed in three independent experiments and the means and standard errors were calculated (***, ANOVA, P< 0.001).
Fig. 4. UPF3B mRNA expression in _SATB2_-deficient cells
(a) Duplex RT-PCR of leukocyte cDNA showing lower levels of UPF3B expression in the SATB2 mutated patient (Pt) compared to those in unaffected female (C1 and C4) or male (C2 and C3) controls. GAPDH was served as a normalizing control gene. Simplex PCR of UPF3B and GAPDH in normal cDNA is shown in lanes 7 and 8. Lane 1; 100-bp DNA ladder. (b) UPF3B mRNA expression by qRT-PCR in leukocyte cDNA of our SATB2 mutated patient is significantly decreased compared to unaffected controls. (**, unpaired t-test, _P_=0.0008). (c) HEK293 cells were transfected with two sets (I and II) of siRNAs against SATB2 or a negative siRNA control. SATB2 and UPF3B expression were measured by qRT-PCR and normalized to β-actin expression. Data are represented as means ± standard deviation (Two sample unequal variant t-test, **P< 0.01). (d) mRNA was extracted from heterozygous and homozygous mutant Satb2 knockout mouse littermate embryonic kidneys (age E18.5). Both Satb2 and Upf3b expression, tested by qRT-PCR, was significantly decreased in mutant (−/−) kidneys compared to that of heterozygous (+/−) littermates. Experiments were performed three times in triplicate. Data are represented as means ± standard deviation. (Two sample unequal variant t-test, *P<0.05, ***P< 0.001).
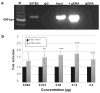
Fig. 5. SATB2 protein binds and activates the UPF3B promoter
(a) Chromatin fragments of Wt SATB2/_myc_-His transfected HEK293 cells were affinity-purified by either a SATB2 antibody (SATB2, lane 2), or a mouse IgG antibody as a negative control (IgG, lane 3), followed by PCR amplification with primers located in the UPF3B promoter. PCR fragments were run on an agarose gel and visualized with ethidium bromide. Input (lane 4) represents PCR products of bulk chromatin extracts (before immunoprecipitation). Positive (+ control gDNA, lane 5) and negative (− control gDNA, lane 6) controls of PCR preparation were included. M (lane 1):100-bp DNA ladder. (b) Firefly luciferase activities from the UPF3B promoter reporter plasmid, normalized to Renilla luciferase activity, were measured with co-expression of increasing amounts of the Wt SATB2/_myc_-His plasmid (gray bars) as indicated. Expression is reported as fold-activation of the SATB2/_myc_-His transfected cells relative to that of the empty pcDNA3.1/_myc_-His vector transfected cells (black bars). Experiments were performed three times in triplicate. Data are represented as means ± standard deviation. (ANOVA, *P<0.05, **P< 0.01, ***P< 0.001).
Similar articles
- Osteoblast-specific transcription factor Osterix (Osx) is an upstream regulator of Satb2 during bone formation.
Tang W, Li Y, Osimiri L, Zhang C. Tang W, et al. J Biol Chem. 2011 Sep 23;286(38):32995-3002. doi: 10.1074/jbc.M111.244236. Epub 2011 Aug 2. J Biol Chem. 2011. PMID: 21828043 Free PMC article. - [Study progress of special AT-rich sequence binding protein 2].
Qian YY, Wang HJ, Ma D. Qian YY, et al. Yi Chuan. 2011 Sep;33(9):947-52. doi: 10.3724/sp.j.1005.2011.00947. Yi Chuan. 2011. PMID: 21951795 Review. Chinese. - Abnormalities in pharyngeal arch-derived structures in SATB2-associated syndrome.
Zarate YA, Bosanko K, Derar N, Fish JL. Zarate YA, et al. Clin Genet. 2024 Aug;106(2):209-213. doi: 10.1111/cge.14540. Epub 2024 May 1. Clin Genet. 2024. PMID: 38693682 - Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects.
Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowanapanja P, Gahl WA, Shotelersuk V. Leoyklang P, et al. Hum Mutat. 2007 Jul;28(7):732-8. doi: 10.1002/humu.20515. Hum Mutat. 2007. PMID: 17377962 - Expanding the phenotype of UPF3B-related disorder: Case reports and literature review.
Romano F, Haanpää MK, Pomianowski P, Peraino AR, Pollard JR, Di Feo MF, Traverso M, Severino M, Derchi M, Henzen E, Zara F, Faravelli F, Capra V, Scala M. Romano F, et al. Am J Med Genet A. 2024 Jun;194(6):e63534. doi: 10.1002/ajmg.a.63534. Epub 2024 Feb 6. Am J Med Genet A. 2024. PMID: 38318947 Review.
Cited by
- Satb2 regulates proliferation and nuclear integrity of pre-osteoblasts.
Dowrey T, Schwager EE, Duong J, Merkuri F, Zarate YA, Fish JL. Dowrey T, et al. Bone. 2019 Oct;127:488-498. doi: 10.1016/j.bone.2019.07.017. Epub 2019 Jul 17. Bone. 2019. PMID: 31325654 Free PMC article. - _SATB2_-associated syndrome caused by a novel SATB2 mutation in a Chinese boy: A case report and literature review.
Zhu YY, Sun GL, Yang ZL. Zhu YY, et al. World J Clin Cases. 2021 Jul 26;9(21):6081-6090. doi: 10.12998/wjcc.v9.i21.6081. World J Clin Cases. 2021. PMID: 34368330 Free PMC article. - Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation.
Addison WN, Hall KC, Kokabu S, Matsubara T, Fu MM, Gori F, Baron R. Addison WN, et al. Mol Cell Biol. 2019 Apr 2;39(8):e00447-18. doi: 10.1128/MCB.00447-18. Print 2019 Apr 15. Mol Cell Biol. 2019. PMID: 30692273 Free PMC article. - Specific behavioural phenotype and secondary cognitive decline as a result of an 8.6 Mb deletion of 2q32.2q33.1.
Gregoric Kumperscak H, Krgovic D, Vokac NK. Gregoric Kumperscak H, et al. J Int Med Res. 2016 Apr;44(2):395-402. doi: 10.1177/0300060515595651. Epub 2016 Jan 25. J Int Med Res. 2016. PMID: 26811410 Free PMC article. - Increased bone turnover, osteoporosis, progressive tibial bowing, fractures, and scoliosis in a patient with a final-exon SATB2 frameshift mutation.
Boone PM, Chan YM, Hunter JV, Pottkotter LE, Davino NA, Yang Y, Beuten J, Bacino CA. Boone PM, et al. Am J Med Genet A. 2016 Nov;170(11):3028-3032. doi: 10.1002/ajmg.a.37847. Epub 2016 Jul 13. Am J Med Genet A. 2016. PMID: 27409069 Free PMC article.
References
- Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Germain G, Gochman P, Long R, Rapoport JL, Rouleau GA. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–239. - PMC - PubMed
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases