MicroRNA expression differentiates squamous epithelium from Barrett's esophagus and esophageal cancer - PubMed (original) (raw)
MicroRNA expression differentiates squamous epithelium from Barrett's esophagus and esophageal cancer
Katherine S Garman et al. Dig Dis Sci. 2013 Nov.
Abstract
Background: Current strategies fail to identify most patients with esophageal adenocarcinoma (EAC) before the disease becomes advanced and incurable. Given the dismal prognosis associated with EAC, improvements in detection of early-stage esophageal neoplasia are needed.
Aim: We sought to assess whether differential expression of microRNAs could discriminate between squamous epithelium, Barrett's esophagus (BE), and EAC.
Methods: We analyzed microRNA expression in a discovery cohort of human endoscopic biopsy samples from 36 patients representing normal squamous esophagus (n = 11), BE (n = 14), and high-grade dysplasia/EAC (n = 11). RNA was assessed using microarrays representing 847 human microRNAs followed by quantitative real-time polymerase chain reaction (qRT-PCR) verification of nine microRNAs. In a second cohort (n = 18), qRT-PCR validation of five miRNAs was performed. Expression of 59 microRNAs associated with BE/EAC in the literature was assessed in our training cohort. Known esophageal cell lines were used to compare miRNA expression to tissue miRNAs.
Results: After controlling for multiple comparisons, we found 34 miRNAs differentially expressed between squamous esophagus and BE/EAC by microarray analysis. However, miRNA expression did not reliably differentiate non-dysplastic BE from EAC. In the validation cohort, all five microRNAs selected for qRT-PCR validation differentiated between squamous samples and BE/EAC. Microarray results supported 14 of the previously reported microRNAs associated with BE/EAC in the literature. Cell lines did not generally reflect miRNA expression found in vivo.
Conclusions: These data indicate that miRNAs differ between squamous esophageal epithelium and BE/EAC, but do not distinguish between BE and EAC. We suggest prospective evaluation of miRNAs in patients at high risk for EAC.
Figures
Figure 1
This flow diagram represents the steps in the analysis using the discovery cohort and a separate validation cohort. Several steps we involved as the initial set of 847 miRNA probe sets from the microarray was narrowed to a list of five miRNAs for validation.
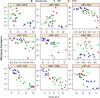
Figure 2
In this figure, the vertical axis represents expression by microarray analysis and the horizontal access represents the delta CT values for each of the nine miRNA selected for qRT-PCR verification of microarray results. This figure provides information on direction of expression by tissue type for both the microarray and PCR platforms. Blue represents squamous samples, green represent BE, while red represents EAC. miR-889, miR-487b, miR-30b and miR-21 increased in BE compared to squamous. miR-27a, miR-92a, miR-27b, miR-193b and miR-149 decreased in BE compared to squamous. The figure also demonstrates concordance between microarray and qRT-PCR results (high concordance appears as a diagonal line). In general, the miRNA expression differentiated squamous tissue from BE/EAC, but not BE from EAC.
Similar articles
- A Systematic Review of Esophageal MicroRNA Markers for Diagnosis and Monitoring of Barrett's Esophagus.
Mallick R, Patnaik SK, Wani S, Bansal A. Mallick R, et al. Dig Dis Sci. 2016 Apr;61(4):1039-50. doi: 10.1007/s10620-015-3959-3. Epub 2015 Nov 14. Dig Dis Sci. 2016. PMID: 26572780 Review. - MicroRNA profile in neosquamous esophageal mucosa following ablation of Barrett's esophagus.
Sreedharan L, Mayne GC, Watson DI, Bright T, Lord RV, Ansar A, Wang T, Kist J, Astill DS, Hussey DJ. Sreedharan L, et al. World J Gastroenterol. 2017 Aug 14;23(30):5508-5518. doi: 10.3748/wjg.v23.i30.5508. World J Gastroenterol. 2017. PMID: 28852310 Free PMC article. - Differential MicroRNA Signatures in the Pathogenesis of Barrett's Esophagus.
Craig MP, Rajakaruna S, Paliy O, Sajjad M, Madhavan S, Reddy N, Zhang J, Bottomley M, Agrawal S, Kadakia MP. Craig MP, et al. Clin Transl Gastroenterol. 2020 Jan;11(1):e00125. doi: 10.14309/ctg.0000000000000125. Clin Transl Gastroenterol. 2020. PMID: 31934893 Free PMC article. - Dynamic changes in microRNA expression profiles reflect progression of Barrett's esophagus to esophageal adenocarcinoma.
Slaby O, Srovnal J, Radova L, Gregar J, Juracek J, Luzna P, Svoboda M, Hajduch M, Ehrmann J. Slaby O, et al. Carcinogenesis. 2015 May;36(5):521-7. doi: 10.1093/carcin/bgv023. Epub 2015 Mar 16. Carcinogenesis. 2015. PMID: 25784377 Clinical Trial. - A review of the current understanding and clinical utility of miRNAs in esophageal cancer.
Sakai NS, Samia-Aly E, Barbera M, Fitzgerald RC. Sakai NS, et al. Semin Cancer Biol. 2013 Dec;23(6 Pt B):512-21. doi: 10.1016/j.semcancer.2013.08.005. Epub 2013 Sep 3. Semin Cancer Biol. 2013. PMID: 24013023 Review.
Cited by
- ESOMIR: a curated database of biomarker genes and miRNAs associated with esophageal cancer.
Sindhoo A, Sipy S, Khan A, Selvaraj G, Alshammari A, Casida ME, Wei DQ. Sindhoo A, et al. Database (Oxford). 2023 Oct 10;2023:baad063. doi: 10.1093/database/baad063. Database (Oxford). 2023. PMID: 37815872 Free PMC article. - MicroRNA Expression can be a Promising Strategy for the Detection of Barrett's Esophagus: A Pilot Study.
Bansal A, Hong X, Lee IH, Krishnadath KK, Mathur SC, Gunewardena S, Rastogi A, Sharma P, Christenson LK. Bansal A, et al. Clin Transl Gastroenterol. 2014 Dec 11;5(12):e65. doi: 10.1038/ctg.2014.17. Clin Transl Gastroenterol. 2014. PMID: 25502391 Free PMC article. - Evaluation of miRNA-196a2 and apoptosis-related target genes: ANXA1, DFFA and PDCD4 expression in gastrointestinal cancer patients: A pilot study.
Fawzy MS, Toraih EA, Ibrahiem A, Abdeldayem H, Mohamed AO, Abdel-Daim MM. Fawzy MS, et al. PLoS One. 2017 Nov 1;12(11):e0187310. doi: 10.1371/journal.pone.0187310. eCollection 2017. PLoS One. 2017. PMID: 29091952 Free PMC article. - MiRNA-Related SNPs and Risk of Esophageal Adenocarcinoma and Barrett's Esophagus: Post Genome-Wide Association Analysis in the BEACON Consortium.
Buas MF, Onstad L, Levine DM, Risch HA, Chow WH, Liu G, Fitzgerald RC, Bernstein L, Ye W, Bird NC, Romero Y, Casson AG, Corley DA, Shaheen NJ, Wu AH, Gammon MD, Reid BJ, Hardie LJ, Peters U, Whiteman DC, Vaughan TL. Buas MF, et al. PLoS One. 2015 Jun 3;10(6):e0128617. doi: 10.1371/journal.pone.0128617. eCollection 2015. PLoS One. 2015. PMID: 26039359 Free PMC article. - A Systematic Review of Esophageal MicroRNA Markers for Diagnosis and Monitoring of Barrett's Esophagus.
Mallick R, Patnaik SK, Wani S, Bansal A. Mallick R, et al. Dig Dis Sci. 2016 Apr;61(4):1039-50. doi: 10.1007/s10620-015-3959-3. Epub 2015 Nov 14. Dig Dis Sci. 2016. PMID: 26572780 Review.
References
- Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett's esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26–33. - PubMed
- Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med. 2011;365:1375–1383. - PubMed
- Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association medical position statement on the management of Barrett's esophagus. Gastroenterology. 2011;140:1084–1091. - PubMed
- National Cancer Institute. [Accessed March 14, 2012, 2012];SEER Cancer Statistics Review. 2011 Available at: http://seer.cancer.gov/csr/1975_2008/.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical