Differential splicing across immune system lineages - PubMed (original) (raw)
. 2013 Aug 27;110(35):14324-9.
doi: 10.1073/pnas.1311839110. Epub 2013 Aug 9.
Collaborators, Affiliations
- PMID: 23934048
- PMCID: PMC3761616
- DOI: 10.1073/pnas.1311839110
Differential splicing across immune system lineages
Ayla Ergun et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Alternative splicing (AS) allows increased diversity and orthogonal regulation of the transcriptional products of mammalian genomes. To assess the distribution and variation of alternative splicing across cell lineages of the immune system, we comprehensively analyzed RNA sequencing and microarray data generated by the Immunological Genome Project Consortium. AS is pervasive: 60% of genes showed frequent AS isoforms in T or B lymphocytes, with 7,599 previously unreported isoforms. Distinct cell specificity was observed, with differential exon skipping in 5% of genes otherwise coexpressed in both B and T cells. The distribution of isoforms was mostly all or none, suggesting on/off switching as a frequent mode of AS regulation in lymphocytes. From the identification of differential exon use in the microarray data, clustering of exon inclusion/exclusion patterns across all Immunological Genome Project cell types showed that ∼70% of AS exons are distributed along a common pattern linked to lineage differentiation and cell cycling. Other AS events distinguished myeloid from lymphoid cells or affected only a small set of exons without clear lineage specificity (e.g., Ptprc). Computational analysis predicted specific associations between AS exons and splicing regulators, which were verified by detection of the hnRPLL/Ptprc connection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
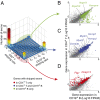
Characterization of splicing junctions from RNA-seq data in CD4+ T and CD19+ B cells. (A) Number (and percent) of reads falling into each of the categories separated by junctions affecting or not affecting the protein-coding sequence (CDS). (B) Number (and percent) of reads affecting the first (denoting an alternative transcriptional start site) or last (denoting alternative polyadenylation) exons in transcripts. (C) Genes with instances of multiple acceptors for a single splice donor and vice versa. (D) Examples of such multiple acceptor or donor sites (Shisa5 and Tcf3) with the number of reads. (E) Alternative acceptor in the sixth exon of Foxp3, which encodes an _N_-truncated protein.
Fig. 2.
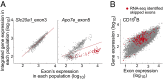
Relative abundance of skipped exons in CD4+ T and CD19+ B cells. (A) Skipping ratio is defined as the number of RNA-seq reads in which an exon is skipped relative to the total number of reads (estimated from neighboring exons reads), and it is plotted for B and T lymphocytes. (B–D) Integrated gene-level expression of CD4+ T and CD19+ B cells, highlighting genes with (C) exons equally skipped in both cells (blue) or (B and D) exons exclusively skipped in B or T cells, respectively.
Fig. 3.
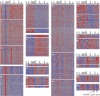
Alternative splicing revealed by feature-level analysis of ST1.0 microarray data. (A) Examples showing the expression of a single exon vs. expression of the corresponding gene across all ImmGen cell types; red highlights (Right) denote DC lineage cells. (B) Plot of every exon’s expression vs. the integrated expression of the corresponding gene in one cell type (here, CD19+ B cells). Exons found to be skipped in the RNA-seq junction data are highlighted (skipping ratio ≥ 0.5).
Fig. 4.
Distribution of exon inclusion/exclusion patterns across all cell types. Optimized _k_-means clustering of 4,321 flagged exons across the ImmGen cell types partitioned exon distributions into 28 clusters. Inclusion/exclusion ratios (exon/gene expression ratios) are shown (
Dataset S4
has a full listing). Clusters 25–28 (1,447 exons, 33.5% of exons considered) did not follow any strong or reproducible pattern, reflecting unique, rare patterns or noise.
Fig. 5.
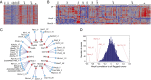
Relation between exon differential use and expression of splicing regulators. Expression of splicing regulators across the ImmGen populations; (A) 27 regulators have distinct patterns of expression, and (B) 138 splicing regulators cluster around one pattern of expression. (C) Significant associations between splicing regulators and inclusion/exclusion of individual exons. (D) Correlation between Hnrpll expression and inclusion/exclusion ratios of all 4,321 flagged exons.
Similar articles
- Rbm24 Regulates Alternative Splicing Switch in Embryonic Stem Cell Cardiac Lineage Differentiation.
Zhang T, Lin Y, Liu J, Zhang ZG, Fu W, Guo LY, Pan L, Kong X, Zhang MK, Lu YH, Huang ZR, Xie Q, Li WH, Xu XQ. Zhang T, et al. Stem Cells. 2016 Jul;34(7):1776-89. doi: 10.1002/stem.2366. Epub 2016 Mar 28. Stem Cells. 2016. PMID: 26990106 - Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes.
Grigoryev YA, Kurian SM, Nakorchevskiy AA, Burke JP, Campbell D, Head SR, Deng J, Kantor AB, Yates JR 3rd, Salomon DR. Grigoryev YA, et al. PLoS One. 2009 Nov 19;4(11):e7906. doi: 10.1371/journal.pone.0007906. PLoS One. 2009. PMID: 19936255 Free PMC article. - SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation.
Lemaire R, Winne A, Sarkissian M, Lafyatis R. Lemaire R, et al. Eur J Immunol. 1999 Mar;29(3):823-37. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. Eur J Immunol. 1999. PMID: 10092085 - Role of RNA Alternative Splicing in T Cell Function and Disease.
Banerjee S, Galarza-Muñoz G, Garcia-Blanco MA. Banerjee S, et al. Genes (Basel). 2023 Sep 30;14(10):1896. doi: 10.3390/genes14101896. Genes (Basel). 2023. PMID: 37895245 Free PMC article. Review.
Cited by
- Mapping the transcriptomics landscape of post-traumatic stress disorder symptom dimensions in World Trade Center responders.
Kuan PF, Yang X, Ren X, Che C, Waszczuk M, Kotov R, Clouston S, Singh PK, Glenn ST, Gomez EC, Wang J, Bromet E, Luft BJ. Kuan PF, et al. Transl Psychiatry. 2021 May 24;11(1):310. doi: 10.1038/s41398-021-01431-6. Transl Psychiatry. 2021. PMID: 34031375 Free PMC article. - Derivation of splice junction-specific antibodies using a unique hapten targeting strategy and directed evolution.
Fuller EP, O'Neill RJ, Weiner MP. Fuller EP, et al. N Biotechnol. 2022 Nov 25;71:1-10. doi: 10.1016/j.nbt.2022.06.003. Epub 2022 Jun 22. N Biotechnol. 2022. PMID: 35750288 Free PMC article. - Heritability and genome-wide association study of vaccine-induced immune response in Beagles: A pilot study.
Blake JM, Thompson J, HogenEsch H, Ekenstedt KJ. Blake JM, et al. Vaccine. 2024 Apr 30;42(12):3099-3106. doi: 10.1016/j.vaccine.2024.03.076. Epub 2024 Apr 10. Vaccine. 2024. PMID: 38604911 Free PMC article. - Genomics of alternative splicing: evolution, development and pathophysiology.
Gamazon ER, Stranger BE. Gamazon ER, et al. Hum Genet. 2014 Jun;133(6):679-87. doi: 10.1007/s00439-013-1411-3. Epub 2014 Jan 1. Hum Genet. 2014. PMID: 24378600 Review.
References
- Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–2144. - PubMed
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30(1):13–19. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous