A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs - PubMed (original) (raw)
A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs
Asma Hatoum-Aslan et al. J Biol Chem. 2013.
Abstract
Small RNAs undergo maturation events that precisely determine the length and structure required for their function. CRISPRs (clustered regularly interspaced short palindromic repeats) encode small RNAs (crRNAs) that together with CRISPR-associated (cas) genes constitute a sequence-specific prokaryotic immune system for anti-viral and anti-plasmid defense. crRNAs are subject to multiple processing events during their biogenesis, and little is known about the mechanism of the final maturation step. We show that in the Staphylococcus epidermidis type III CRISPR-Cas system, mature crRNAs are measured in a Cas10·Csm ribonucleoprotein complex to yield discrete lengths that differ by 6-nucleotide increments. We looked for mutants that impact this crRNA size pattern and found that an alanine substitution of a conserved aspartate residue of Csm3 eliminates the 6-nucleotide increments in the length of crRNAs. In vitro, recombinant Csm3 binds RNA molecules at multiple sites, producing gel-shift patterns that suggest that each protein binds 6 nucleotides of substrate. In vivo, changes in the levels of Csm3 modulate the crRNA size distribution without disrupting the 6-nucleotide periodicity. Our data support a model in which multiple Csm3 molecules within the Cas10·Csm complex bind the crRNA with a 6-nucleotide periodicity to function as a ruler that measures the extent of crRNA maturation.
Keywords: Adaptive Immunity; Antibiotic Resistance; Bacterial Conjugation; Bacterial Genetics; CRISPR; Microbiology; Small RNA; Staphylococcus.
Figures
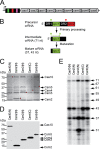
FIGURE 1.
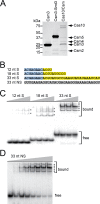
Mature crRNAs are measured in a Cas10·Csm complex. A, organization of the type III-A CRISPR system in S. epidermidis RP62A. This system contains 9 CRISPR-associated (cas and csm) genes, 4 direct repeats (black boxes), and 3 spacers (colored boxes), the first of which targets the nickase gene in staphylococcal conjugative plasmids. B, a ruler mechanism determines the length of mature crRNAs. Transcription of the repeat-spacer array generates a precursor crRNA that is subject to two cleavage events: primary processing within repeats to yield ∼71-nt intermediates (filled triangles), and maturation through trimming of the 3′ end of the intermediate (empty triangles). A ruler mechanism anchored at the primary processing site determines the extension of maturation to generate 37- and 43-nt-long mature crRNA. C, the type III-A Cas10·Csm complex. His6 tags were placed on the indicated (N or C) terminus of each of the genes involved in crRNA biogenesis. Constructs were expressed in S. epidermidis LM1680, and whole cell lysates were subject to Ni2+ affinity chromatography. Complexes were resolved by SDS-PAGE and visualized using coomassie G-250 staining. Red asterisks indicate each of the tagged species. D, each tagged Cas10·Csm subunit was visualized by Western blot of cell extracts using Ni2+-HRP. E, RNA was extracted from each of the His6- Cas10·Csm complexes, radiolabeled at the 5′ end, and resolved using denaturing PAGE.
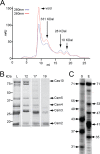
FIGURE 2.
Purification of the Cas10·Csm complex from E. coli. A, elution profile of the type III-A Cas10·Csm (Csm2H6N) complex extracted from E. coli. Whole cell lysates were subject to Ni2+ affinity chromatography, followed by size exclusion with a Superdex 200 column. UV absorbances at _A_260 (red) and _A_280 (blue) are shown in miliabsorbance units (mAU) as a function of elution volume. Molecular weight estimates for each peak are indicated. B, protein content for the load (L) and indicated 1-ml fractions (12, 17, and 19) resolved by SDS-PAGE and visualized using Coomassie G-250 staining. C, crRNAs associated with Cas10·Csm (Csm2H6N) complexes extracted from S. epidermidis (S) and E. coli (E) are shown. crRNAs extracted from purified complexes were radio-labeled on their 5′ end and resolved using denaturing PAGE.
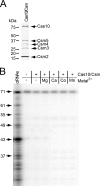
FIGURE 3.
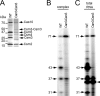
The Cas10·Csm complex does not contain maturation activity in vitro. A, Cas10·Csm (Csm2H6N) purified from a p_crispr_/Δ_cas6_ construct in S. epidermidis LM1680. Whole cell lysates were subjected to Ni2+ affinity chromatography. The Δ_cas6_ construct was used to ensure the absence of mature crRNAs preloaded into the complex. B, a 65-nt spc1 crRNA substrate was 5′ end-labeled using polynucleotide kinase, PAGE-purified, and combined with the complex (500 pmol) in the presence of EDTA or various metals as indicated. The reaction mixture was incubated at 37 °C for 20 min, and RNAs were resolved by denaturing PAGE.
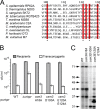
FIGURE 4.
Csm3 modulates the length of mature crRNAs. A, partial Csm3 sequence alignment using BLAST. Highly conserved residues are highlighted in red, mutated residues are indicated with asterisks. B, mutations in conserved Csm3 residues do not affect CRISPR interference against the conjugative plasmid pG0400. S. epidermidis LM1680 strains harboring wild type and mutant p_crispr_ plasmids with the indicated alanine substitutions in Csm3 and encoding for His6-Csm2 were used as recipients for the transfer of pG0400. Conjugation was carried out in duplicate; the values (in cfu/ml; mean ± S.D.) obtained for recipients and transconjugants are shown. C, wild type and mutant Cas10·Csm (Csm2H6N) complexes were purified from S. epidermidis, and crRNAs were extracted, radiolabeled on their 5′ end, and resolved using denaturing PAGE. Whereas most mutations do not affect severely the crRNA size distribution, the D100A substitution leads to the elimination of all crRNAs except the 31-nt species.
FIGURE 5.
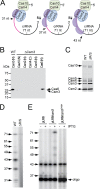
Multiple copies of Csm3 bind crRNAs with a 6-nucleotide periodicity. A, Csm3 was cloned into the pET28b vector in frame with an N-terminal His10 tag and the Smt3 peptide. The construct was expressed in E. coli and whole cell lysates were subject to Ni2+ affinity chromatography, followed by dialysis with SUMO protease, which cleaves the His10-Smt3 tag. A second Ni2+ affinity purification was used to separate the tag. Csm3 preps before and after cleavage of the tag (Smt3-Csm3 and Csm3, respectively) were resolved by SDS-PAGE and visualized using Coomassie G-250 staining. B, RNA substrates used for Csm3 binding assays. Specific substrates (S) are identical to spc1 crRNAs, harboring the repeat sequence corresponding to the 5′, 8-nt tag (blue) and various lengths of spacer sequence (yellow). The nonspecific substrate (NS, gray) harbors no resemblance to a crRNA. C, Csm3 binding to the spc1 crRNA substrates of different lengths is shown. Trace amounts of end-labeled substrate were incubated with increasing amounts of Csm3 (0, 256, 320, and 384 pmol). Bound and free substrates are indicated. D, Csm3 binding to a nonspecific substrate. Trace amounts of end-labeled substrate were incubated with increasing amounts of Csm3 (0, 8, 16, 32, 64, 128, 185, and 256 pmol). Bound and free substrates are indicated.
FIGURE 6.
Csm3 protects 6 nt of the crRNAs during maturation. A, purification of Cas10·Csm complexes containing a Csm3-Csm3 dimer. The complex was purified from a strain expressing His6-Csm2 and a dimer consisting of two Csm3 proteins tethered through a flexible (GGGGS)3 linker (Csm3-Csm3) using Ni2+ affinity chromatography. B and C, total crRNAs were extracted from the indicated Cas10·Csm complexes (B), or spc1 crRNAs were captured from total RNA extracts of indicated strains using a biotinylated probe antisense to spc1 (C, probe marked with a black arrowhead). crRNAs were radiolabeled at the 5′ end and resolved using denaturing PAGE.
FIGURE 7.
Csm3 levels determine the extent of crRNA maturation. A, a model for crRNA maturation in which Csm3 protects the 3′ end of crRNAs acting as a 6-nt ruler within the Cas10·Csm complex to dictate the extent of maturation. B, a Western blot is shown using Ni2+-HRP to probe for each of the indicated His-tagged species in a Δ_csm3_ background. His6 tags were placed on p_crispr_/Δ_csm3_ on each remaining member of the complex (as indicated), and also in Cas6, and constructs were expressed in S. epidermidis LM1680. Whole cell lysates of each strain were subject to Ni2+ affinity chromatography, and extracts were resolved by SDS-PAGE prior to Western blotting. Although Cas6 can be detected in cell extracts, the members of the Cas10·Csm complex cannot. C, SDS-PAGE of a His6·Csm2 complex expressed from a p_crispr_ plasmid lacking the repeat/spacer sequences (Δ_R_/S) and purified using Ni2+ affinity chromatography. D, extraction, labeling, and PAGE separation of RNAs from the Δ_R_/S Cas10·Csm complex. E, Spc1 crRNAs visualized in wild type S. epidermidis in the presence of pLM9, pLM9/csm3, or pLM9/_csm3_D100A. Where indicated, Csm3 overexpression was induced with isopropyl 1-thio-β-
d
-galactopyranoside (IPTG) for 3 h prior to harvesting total RNA. crRNAs were captured from total RNA extracts using a biotinylated probe antisense to spc1, radiolabeled at the 5′ end, and resolved using denaturing PAGE. oligo, oligonucleotide probe.
Similar articles
- Molecular determinants for CRISPR RNA maturation in the Cas10-Csm complex and roles for non-Cas nucleases.
Walker FC, Chou-Zheng L, Dunkle JA, Hatoum-Aslan A. Walker FC, et al. Nucleic Acids Res. 2017 Feb 28;45(4):2112-2123. doi: 10.1093/nar/gkw891. Nucleic Acids Res. 2017. PMID: 28204542 Free PMC article. - Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system.
Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Hatoum-Aslan A, et al. J Bacteriol. 2014 Jan;196(2):310-7. doi: 10.1128/JB.01130-13. Epub 2013 Nov 1. J Bacteriol. 2014. PMID: 24187086 Free PMC article. - Crystal Structures of Csm2 and Csm3 in the Type III-A CRISPR-Cas Effector Complex.
Takeshita D, Sato M, Inanaga H, Numata T. Takeshita D, et al. J Mol Biol. 2019 Feb 15;431(4):748-763. doi: 10.1016/j.jmb.2019.01.009. Epub 2019 Jan 11. J Mol Biol. 2019. PMID: 30639408 - Approaches to study CRISPR RNA biogenesis and the key players involved.
Behler J, Hess WR. Behler J, et al. Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review. - Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems.
Richter C, Chang JT, Fineran PC. Richter C, et al. Viruses. 2012 Oct 19;4(10):2291-311. doi: 10.3390/v4102291. Viruses. 2012. PMID: 23202464 Free PMC article. Review.
Cited by
- Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus.
Majumdar S, Zhao P, Pfister NT, Compton M, Olson S, Glover CV 3rd, Wells L, Graveley BR, Terns RM, Terns MP. Majumdar S, et al. RNA. 2015 Jun;21(6):1147-58. doi: 10.1261/rna.049130.114. Epub 2015 Apr 22. RNA. 2015. PMID: 25904135 Free PMC article. - Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L. Gleditzsch D, et al. Nucleic Acids Res. 2016 Jul 8;44(12):5872-82. doi: 10.1093/nar/gkw469. Epub 2016 May 23. Nucleic Acids Res. 2016. PMID: 27216815 Free PMC article. - A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity.
Chou-Zheng L, Hatoum-Aslan A. Chou-Zheng L, et al. Elife. 2019 Apr 3;8:e45393. doi: 10.7554/eLife.45393. Elife. 2019. PMID: 30942690 Free PMC article. - A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH. Hagelueken G, et al. Nat Struct Mol Biol. 2015 Jan;22(1):50-56. doi: 10.1038/nsmb.2935. Epub 2014 Dec 15. Nat Struct Mol Biol. 2015. PMID: 25504321 Free PMC article. - The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Paraan M, Nasef M, Chou-Zheng L, Khweis SA, Schoeffler AJ, Hatoum-Aslan A, Stagg SM, Dunkle JA. Paraan M, et al. PLoS One. 2023 Jun 23;18(6):e0287461. doi: 10.1371/journal.pone.0287461. eCollection 2023. PLoS One. 2023. PMID: 37352230 Free PMC article.
References
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 - PubMed
- Garneau J. E., Dupuis M. È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials