Phylogenetic relationships and species delimitation in pinus section trifoliae inferrred from plastid DNA - PubMed (original) (raw)
Phylogenetic relationships and species delimitation in pinus section trifoliae inferrred from plastid DNA
Sergio Hernández-León et al. PLoS One. 2013.
Abstract
Recent diversification followed by secondary contact and hybridization may explain complex patterns of intra- and interspecific morphological and genetic variation in the North American hard pines (Pinus section Trifoliae), a group of approximately 49 tree species distributed in North and Central America and the Caribbean islands. We concatenated five plastid DNA markers for an average of 3.9 individuals per putative species and assessed the suitability of the five regions as DNA bar codes for species identification, species delimitation, and phylogenetic reconstruction. The ycf1 gene accounted for the greatest proportion of the alignment (46.9%), the greatest proportion of variable sites (74.9%), and the most unique sequences (75 haplotypes). Phylogenetic analysis recovered clades corresponding to subsections Australes, Contortae, and Ponderosae. Sequences for 23 of the 49 species were monophyletic and sequences for another 9 species were paraphyletic. Morphologically similar species within subsections usually grouped together, but there were exceptions consistent with incomplete lineage sorting or introgression. Bayesian relaxed molecular clock analyses indicated that all three subsections diversified relatively recently during the Miocene. The general mixed Yule-coalescent method gave a mixed model estimate of only 22 or 23 evolutionary entities for the plastid sequences, which corresponds to less than half the 49 species recognized based on morphological species assignments. Including more unique haplotypes per species may result in higher estimates, but low mutation rates, recent diversification, and large effective population sizes may limit the effectiveness of this method to detect evolutionary entities.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
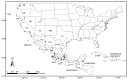
Figure 1. Geographic distribution of individual samples of Pinus section Trifoliae.
Three of the 191 individuals are of unknown provenance.
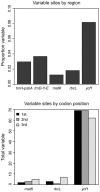
Figure 2. Variation of the plastid markers in Pinus section Trifoliae.
Above: The proportion of variable sites for each of the five markers. Below: the total number of variable sites per codon position for the three protein coding regions.
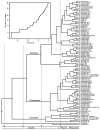
Figure 3. Maximum likelihood tree.
Bootstrap values greater than 50% are shown at branches. A. Pinus subsection Australes. Images from top to bottom are of P . lumholtzii, P . herrerae, P . oocarpa, P . chihuahuana, and P . attenuata. B. Pinus subsections Ponderosae and Contortae. Images from top to bottom are P . scopulorum, P . douglasiana, P . pseudostrobus var. apulcensis, P . coulteri, and P . contorta. The branch leading to the outgroups has been truncated.
Figure 4. Chronogram for Pinus section Trifoliae using three calibration points and lineage-through-time plot.
Chronogram for section Trifoliae. The outgroup has been removed. The two secondary calibration points inferred from a previous relaxed molecular clock are indicated with an asterisk (*), and a fossil calibration point attributed to the California closed-cone pines is indicated with a double asterisk (**). The corresponding lineage-through-time plot is given on the upper left. The transition from speciation to coalescence branching processes gave nonsignificant test results.
Similar articles
- Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes.
Gernandt DS, Aguirre Dugua X, Vázquez-Lobo A, Willyard A, Moreno Letelier A, Pérez de la Rosa JA, Piñero D, Liston A. Gernandt DS, et al. Am J Bot. 2018 Apr;105(4):711-725. doi: 10.1002/ajb2.1052. Epub 2018 Apr 23. Am J Bot. 2018. PMID: 29683492 - Phylogeography of Pinus subsection Australes in the Caribbean Basin.
Jardón-Barbolla L, Delgado-Valerio P, Geada-López G, Vázquez-Lobo A, Piñero D. Jardón-Barbolla L, et al. Ann Bot. 2011 Feb;107(2):229-41. doi: 10.1093/aob/mcq232. Epub 2010 Nov 29. Ann Bot. 2011. PMID: 21118838 Free PMC article. - Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): implications for Northern Hemisphere biogeography.
Hao ZZ, Liu YY, Nazaire M, Wei XX, Wang XQ. Hao ZZ, et al. Mol Phylogenet Evol. 2015 Jun;87:65-79. doi: 10.1016/j.ympev.2015.03.013. Epub 2015 Mar 20. Mol Phylogenet Evol. 2015. PMID: 25800283 - Coalescent-based species delimitation in North American pinyon pines using low-copy nuclear genes and plastomes.
Montes JR, Peláez P, Moreno-Letelier A, Gernandt DS. Montes JR, et al. Am J Bot. 2022 May;109(5):706-726. doi: 10.1002/ajb2.1847. Epub 2022 May 8. Am J Bot. 2022. PMID: 35526278 Free PMC article. - Pinus ponderosa: A checkered past obscured four species.
Willyard A, Gernandt DS, Potter K, Hipkins V, Marquardt P, Mahalovich MF, Langer SK, Telewski FW, Cooper B, Douglas C, Finch K, Karemera HH, Lefler J, Lea P, Wofford A. Willyard A, et al. Am J Bot. 2017 Jan;104(1):161-181. doi: 10.3732/ajb.1600336. Epub 2016 Dec 28. Am J Bot. 2017. PMID: 28031167
Cited by
- Morphological and niche divergence of pinyon pines.
Ortiz-Medrano A, Scantlebury DP, Vázquez-Lobo A, Mastretta-Yanes A, Piñero D. Ortiz-Medrano A, et al. Ecol Evol. 2016 Mar 23;6(9):2886-96. doi: 10.1002/ece3.1994. eCollection 2016 May. Ecol Evol. 2016. PMID: 27092235 Free PMC article. - Tall Pinus luzmariae trees with genes from P. herrerae.
Wehenkel C, Mariscal-Lucero SDR, González-Elizondo MS, Aguirre-Galindo VA, Fladung M, López-Sánchez CA. Wehenkel C, et al. PeerJ. 2020 Feb 26;8:e8648. doi: 10.7717/peerj.8648. eCollection 2020. PeerJ. 2020. PMID: 32149029 Free PMC article. - New Insight into Taxonomy of European Mountain Pines, Pinus mugo Complex, Based on Complete Chloroplast Genomes Sequencing.
Sokołowska J, Fuchs H, Celiński K. Sokołowska J, et al. Plants (Basel). 2021 Jun 29;10(7):1331. doi: 10.3390/plants10071331. Plants (Basel). 2021. PMID: 34209970 Free PMC article. - Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines.
Jin WT, Gernandt DS, Wehenkel C, Xia XM, Wei XX, Wang XQ. Jin WT, et al. Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2022302118. doi: 10.1073/pnas.2022302118. Proc Natl Acad Sci U S A. 2021. PMID: 33941644 Free PMC article. - Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros.
Li W, Liu Y, Yang Y, Xie X, Lu Y, Yang Z, Jin X, Dong W, Suo Z. Li W, et al. BMC Plant Biol. 2018 Sep 26;18(1):210. doi: 10.1186/s12870-018-1421-3. BMC Plant Biol. 2018. PMID: 30257644 Free PMC article.
References
- Hudson RR (1990) Gene genealogies and the coalescent process. Oxf Surv Evol Biol 7: 1-44.
- Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants 5: 65-84.
- Maddison W (1995) Phylogenetic histories within and among species. In: Hoch PC, Stevenson AG. Experimental and molecular approaches to plant biosystematics Monographs in systematics. St. Louis, Missouri: Botanical Garden; pp. 273-287.
- Maddison WP (1997) Gene trees in species trees. Syst Biol 46: 523-536. doi:10.1093/sysbio/46.3.523. - DOI
- Rosenberg NA (2003) The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphily, and polyphyly in a coalescent model. Evolution 57: 1465-1477. doi:10.1554/03-012. PubMed: 12940352. - DOI - PubMed
MeSH terms
Substances
Grants and funding
This study was supported by the Dirección General de Asuntos del Personal Académico-Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica of the Universidad Nacional Autónoma de México (DGAPA-PAPIIT; IN228209), the United Nations Development Programme, the National Commission for Knowledge and Use of Biodiversity (CONABIO; GE021), and the National Council on Science and Technology (CONACYT; The Barcodes of Life Thematic Network). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources