Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis - PubMed (original) (raw)
Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis
Zhonggang Hou et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Genome engineering in human pluripotent stem cells (hPSCs) holds great promise for biomedical research and regenerative medicine. Recently, an RNA-guided, DNA-cleaving interference pathway from bacteria [the type II clustered, regularly interspaced, short palindromic repeats (CRISPR)-CRISPR-associated (Cas) pathway] has been adapted for use in eukaryotic cells, greatly facilitating genome editing. Only two CRISPR-Cas systems (from Streptococcus pyogenes and Streptococcus thermophilus), each with their own distinct targeting requirements and limitations, have been developed for genome editing thus far. Furthermore, limited information exists about homology-directed repair (HDR)-mediated gene targeting using long donor DNA templates in hPSCs with these systems. Here, using a distinct CRISPR-Cas system from Neisseria meningitidis, we demonstrate efficient targeting of an endogenous gene in three hPSC lines using HDR. The Cas9 RNA-guided endonuclease from N. meningitidis (NmCas9) recognizes a 5'-NNNNGATT-3' protospacer adjacent motif (PAM) different from those recognized by Cas9 proteins from S. pyogenes and S. thermophilus (SpCas9 and StCas9, respectively). Similar to SpCas9, NmCas9 is able to use a single-guide RNA (sgRNA) to direct its activity. Because of its distinct protospacer adjacent motif, the N. meningitidis CRISPR-Cas machinery increases the sequence contexts amenable to RNA-directed genome editing.
Keywords: crRNA; embryonic stem cells; induced pluripotent stem cells; tracrRNA.
Conflict of interest statement
Conflict of interest statement: Northwestern University (Y.Z. and E.J.S.) and Wisconsin Alumni Research Foundation (Z.H. and J.A.T.) have filed a related patent: DNA cleavage and genome editing using Cas9 from Neisseria meningitidis.
Figures
Fig. 1.
Functional NmCas9 can be expressed in mammalian cells. (A) Western blot analysis demonstrates that FLAG-tagged NmCas9 is expressed in 293FT cells. (Lane 1) Untransfected 293FT cells. (Lane 2) 293FT cells transfected with FLAG-tagged NmCas9 expressing plasmid. (Upper) Anti-FLAG tag Western. (Lower) Anti-GAPDH Western as loading control. (B) Design of the crRNA that targets the tdTomato coding sequence. Blue: PAM sequence; orange: crRNA spacer; green: crRNA repeat. (C) The plasmid containing the tdTomato coding sequence (see B) was linearized with NdeI and mixed with different combinations of tracrRNA, crRNA, and cell lysate prepared from NmCas9-expressing 293FT cells. After incubation at 37 °C, DNA was purified and analyzed by agarose gel electrophoresis. The diagram on the right shows the expected cleavage products and their predicted sizes. “N” indicates inclusion of a nonspecific crRNA that does not target tdTomato. (D) Cleavage product (see C) was extracted from the gel and analyzed by Sanger sequencing using the primers indicated in the Right panel. The cleavage site, indicated by the arrow, was inferred from the sequencing.
Fig. 2.
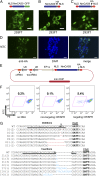
NmCas9 functions in gene disruption in human ES cells. (A_–_C) The localization of NmCas9 with an NLS at the N terminus (A), C terminus (B), or both termini (C) was analyzed by either EGFP fluorescence (A) or anti-HA immunofluorescence (B and C) in 293FT cells. (Scale bars, 20 µm.) (D) The localization of NmCas9 with the double NLS (see C) was analyzed by anti-HA immunofluorescence in hESCs. (Scale bars, 20 µm.) (E) Design of a single plasmid used for gene editing in hESCs. (F) FACS analysis of tdTomato reporter hESC lines after electroporation of the indicated crRNA/tracrRNA/NmCas9 constructs. The number in the plot indicates the percentage of tdTomato-fluorescence-negative cells 5 d after electroporation. (G) Indels introduced by the targeting CRISPR in the tdTomato negative population (see F) were analyzed by targeted PCR amplification and sequencing. The protospacer sequence is underlined. The numbers in parentheses indicate the number of sequenced clones containing that specific indel.
Fig. 3.
Specificity screen of NmCas9 system. (A, Upper) The crRNA sequence targeting tdTomato, with locations of single point mutations (black) in the spacer region of the tdTomato targeting crRNA. (Lower) The efficiency of each mutant at disrupting tdTomato expression. All mutants were tested as described in Fig. 2_F_. The efficiency is defined as percent tdTomato-negative cells (mutant spacer) divided by percent tdTomato-negative cells (wild-type spacer). Error bars: SEM (B, Upper) The locations of different mutant PAMs in the tdTomato sequence. Blue, wild-type PAM; brown, mutant PAM; underlined, spacer sequence. For the bottom-most protospacer, the opposite strand was targeted, and the reverse complement sequence is therefore shown. (Lower) The efficiency of targeting at each site associated with the indicated PAM, as revealed by the loss of tdTomato expression. All targeting experiments were performed as described in Fig. 2_F_. The efficiency is defined as percent tdTomato-negative cells (mutant PAM) divided by percent tdTomato-negative cells (wild-type PAM). Error bars: SEM.
Fig. 4.
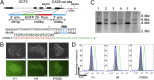
Gene targeting in hESCs using NmCas9. (A) Donor DNA and crRNA design. The mismatch in the first nucleotide of crRNA is to satisfy the requirement of the U6 promoter for a G residue at the transcription start site. (B) Phase-contrast (Upper) and fluorescent (Lower) images of targeted clones from H1, H9, and iPS005 line. (Scale bars, 50 µM.) (C) Southern blot analysis of H1 WT (lane 1), H9 WT (lane 3), and iPS005 WT (lane 5) and targeted clones of H1 (lane 2), H9 (lane 4), and iPS005 (lane 6) line. Genomic DNA was digested with BamHI. The Southern probe is located outside of donor DNA (see A). The wild-type clone should give one band of 4.2 kb and targeted heterozygous clone should give one additional band of 5.6 kb. (D) Targeted clones (see B) were treated with 10 µM SB431542 and 10 ng/mL BMP4 to initiate differentiation. The EGFP signal was analyzed by FACS 3 d after differentiation. Gray, undifferentiated parental cells before targeting; green, targeted cells before differentiation; blue, targeted cells after differentiation.
Comment in
- A variant CRISPR-Cas9 system adds versatility to genome engineering.
Walsh RM, Hochedlinger K. Walsh RM, et al. Proc Natl Acad Sci U S A. 2013 Sep 24;110(39):15514-5. doi: 10.1073/pnas.1314697110. Epub 2013 Sep 6. Proc Natl Acad Sci U S A. 2013. PMID: 24014593 Free PMC article. No abstract available.
Similar articles
- The iCRISPR platform for rapid genome editing in human pluripotent stem cells.
Zhu Z, González F, Huangfu D. Zhu Z, et al. Methods Enzymol. 2014;546:215-50. doi: 10.1016/B978-0-12-801185-0.00011-8. Methods Enzymol. 2014. PMID: 25398343 Free PMC article. - The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells.
Lee CM, Cradick TJ, Bao G. Lee CM, et al. Mol Ther. 2016 Mar;24(3):645-54. doi: 10.1038/mt.2016.8. Epub 2016 Jan 19. Mol Ther. 2016. PMID: 26782639 Free PMC article. - A cut above the rest: targeted genome editing technologies in human pluripotent stem cells.
Li M, Suzuki K, Kim NY, Liu GH, Izpisua Belmonte JC. Li M, et al. J Biol Chem. 2014 Feb 21;289(8):4594-9. doi: 10.1074/jbc.R113.488247. Epub 2013 Dec 20. J Biol Chem. 2014. PMID: 24362028 Free PMC article. Review. - [Efficient genome editing in human pluripotent stem cells through CRISPR/Cas9].
Liu GG, Li S, Wei YD, Zhang YX, Ding QR. Liu GG, et al. Yi Chuan. 2015 Nov;37(11):1167-73. doi: 10.16288/j.yczz.15-240. Yi Chuan. 2015. PMID: 26582531 Chinese. - [CRISPR/Cas system for genome editing in pluripotent stem cells].
Vasil'eva EA, Melino D, Barlev NA. Vasil'eva EA, et al. Tsitologiia. 2015;57(1):19-30. Tsitologiia. 2015. PMID: 25872372 Review. Russian.
Cited by
- Harnessing CRISPR-Cas system diversity for gene editing technologies.
Mckay A, Burgio G. Mckay A, et al. J Biomed Res. 2021 Mar 26;35(2):91-106. doi: 10.7555/JBR.35.20200184. J Biomed Res. 2021. PMID: 33797415 Free PMC article. - Recent Advances in Genome Editing Using CRISPR/Cas9.
Ding Y, Li H, Chen LL, Xie K. Ding Y, et al. Front Plant Sci. 2016 May 24;7:703. doi: 10.3389/fpls.2016.00703. eCollection 2016. Front Plant Sci. 2016. PMID: 27252719 Free PMC article. Review. - Efficient CRISPR/Cas9-Mediated Versatile, Predictable, and Donor-Free Gene Knockout in Human Pluripotent Stem Cells.
Liu Z, Hui Y, Shi L, Chen Z, Xu X, Chi L, Fan B, Fang Y, Liu Y, Ma L, Wang Y, Xiao L, Zhang Q, Jin G, Liu L, Zhang X. Liu Z, et al. Stem Cell Reports. 2016 Sep 13;7(3):496-507. doi: 10.1016/j.stemcr.2016.07.021. Epub 2016 Sep 1. Stem Cell Reports. 2016. PMID: 27594587 Free PMC article. - Research on CRISPR/system in major cancers and its potential in cancer treatments.
Liu Z, Liao Z, Chen Y, Zhou L, Huangting W, Xiao H. Liu Z, et al. Clin Transl Oncol. 2021 Mar;23(3):425-433. doi: 10.1007/s12094-020-02450-3. Epub 2020 Jul 15. Clin Transl Oncol. 2021. PMID: 32671729 Review. - Fluorescent protein biosensors applied to microphysiological systems.
Senutovitch N, Vernetti L, Boltz R, DeBiasio R, Gough A, Taylor DL. Senutovitch N, et al. Exp Biol Med (Maywood). 2015 Jun;240(6):795-808. doi: 10.1177/1535370215584934. Epub 2015 May 19. Exp Biol Med (Maywood). 2015. PMID: 25990438 Free PMC article. Review.
References
- Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. - PubMed
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21(3):319–321. - PubMed
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM093769/GM/NIGMS NIH HHS/United States
- UH2 TR000506/TR/NCATS NIH HHS/United States
- UH3 TR000506/TR/NCATS NIH HHS/United States
- 1UH2TR000506-01/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous