The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges - PubMed (original) (raw)
Review
The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges
Andrew P Hutchins et al. Brief Funct Genomics. 2013 Nov.
Abstract
Inflammation is a fundamental response of the immune system whose successful termination involves the elimination of the invading pathogens, the resolution of inflammation and the repair of the local damaged tissue. In this context, the interleukin 10 (IL-10)-mediated anti-inflammatory response (AIR) represents an essential homeostatic mechanism that controls the degree and duration of inflammation. Here, we review recent work on the mechanistic characterization of the IL-10-mediated AIR on multiple levels: from the cataloguing of the in vivo genomic targets of STAT3 (the transcription factor downstream of IL-10) to the identification of specific co-factors that endow STAT3 with genomic-binding specificity, and how genomic and computational methods are being used to elucidate the regulatory mechanisms of this essential physiological response in macrophages.
Keywords: IL-10; JAK1; STAT3; anti-inflammatory response; bioinformatics; macrophages; transcriptional regulatory modules.
Figures
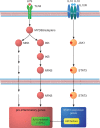
Figure 1:
The IL-10 AIR in the context of inflammation. The TLR-initiated inflammatory response activates a set of well-understood signalling pathways, which ultimately lead to the expression of thousands of pro-inflammatory genes. IL-10 initiates the AIR upon binding to its cognate receptor, where JAK1 phosphorylates STAT3. Neither IL-10, the IL-10R, the Janus kinase or STAT3 can be functionally replaced in this pathway. Upon entering the nucleus, STAT3 activates specific target genes among which the ultimate effectors of the AIR (the ‘AIR factors’) are found. The AIR factors repress the expression of pro-inflammatory genes mostly at the transcriptional level, with only ∼20% of the genes activated by LPS in macrophages eventually being repressed by IL-10. Adapted from Murray [11].
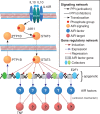
Figure 2:
Characterization of TRMs. (A) The goal of the rTRM method is to reconstruct TRMs by integrating multiple data sources. Starting with a set of experimentally characterized genomic-binding sites (e.g. from ChIP-seq) (1), these sites are investigated for the presence of over-represented TF binding motifs using computational approaches (2). The over-represented motifs are matched to their corresponding TFs, and these are further filtered by expression data from the same cell type (3). Finally, rTRM harnesses the power of PPIs by investigating the entire PPI space from the BioGRID database to build TRMs from the TFs that have survived the previous filtering steps. (B) The genomic-binding patterns of STAT3 encode two distinct modes of regulation: a ‘macrophage-specific’ regulatory mode characterized by a specific set of co-TFs and co-factors (left panel) and a ‘universal’ regulatory mode characterized by a distinct TRM. Whereas the universal mode of STAT3 regulation serves to perpetuate STAT3 signalling and fine-tune the JAK-STAT pathway, the macrophage-specific mode is responsible for the AIR.
Figure 3:
The IL-10/JAK1/STAT3 anti-inflammatory pathway: current state of affairs. The ChIP-seq of STAT3 in IL-10-stimulated macrophages, coupled with RNA-seq, has unveiled the repertoire of genes that are activated by STAT3 during the AIR. Among these the AIR factors shall be found, followed by a detailed characterization of the AIR factor-target gene relationships, and their mechanisms. Another fundamental aspect of the AIR that needs addressing is the role that the epigenetic landscape plays in regulating the AIR, as well as the potential regulation of the AIR by STAT3-induced enzymes, such as PTP1B, a tyrosine phosphatase previously shown to target phospho-STAT3 [56]. An integrated understanding of the AIR factors and their mechanisms of action on their targets, the genomic regulation of the AIR by specific TRMs and by the epigenetic landscape, as well as the cytoplasmic-level regulation of the AIR will provide us with an unprecedented degree of detail on this essential physiological response that we may harness to develop tailored anti-inflammatory therapies.
Similar articles
- Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages.
Hutchins AP, Poulain S, Miranda-Saavedra D. Hutchins AP, et al. Blood. 2012 Mar 29;119(13):e110-9. doi: 10.1182/blood-2011-09-381483. Epub 2012 Feb 9. Blood. 2012. PMID: 22323479 - Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response.
Murray PJ. Murray PJ. Curr Opin Pharmacol. 2006 Aug;6(4):379-86. doi: 10.1016/j.coph.2006.01.010. Epub 2006 May 18. Curr Opin Pharmacol. 2006. PMID: 16713356 Review. - Aryl Hydrocarbon Receptor Promotes IL-10 Expression in Inflammatory Macrophages Through Src-STAT3 Signaling Pathway.
Zhu J, Luo L, Tian L, Yin S, Ma X, Cheng S, Tang W, Yu J, Ma W, Zhou X, Fan X, Yang X, Yan J, Xu X, Lv C, Liang H. Zhu J, et al. Front Immunol. 2018 Sep 19;9:2033. doi: 10.3389/fimmu.2018.02033. eCollection 2018. Front Immunol. 2018. PMID: 30283437 Free PMC article. - Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses.
Hutchins AP, Takahashi Y, Miranda-Saavedra D. Hutchins AP, et al. Sci Rep. 2015 Mar 13;5:9100. doi: 10.1038/srep09100. Sci Rep. 2015. PMID: 25765318 Free PMC article. - The IL-10/STAT3 axis: Contributions to immune tolerance by thymus and peripherally derived regulatory T-cells.
Schmetterer KG, Pickl WF. Schmetterer KG, et al. Eur J Immunol. 2017 Aug;47(8):1256-1265. doi: 10.1002/eji.201646710. Epub 2017 Jul 19. Eur J Immunol. 2017. PMID: 28631311 Review.
Cited by
- Cord blood levels of interleukin-10 decrease in neonates with increased birth weight: novel implications of the cytokine network in early obesity.
Méndez-García LA, Minor-Borrego H, Sánchez-Del Real AL, Aguayo-Guerrero JA, Alvarado-Monroy T, Trejo-Millán F, Rosas-Salinas J, Rizo-Tellez SA, Islas-Andrade S, Briones-Garduño JC, Fragoso JM, Escobedo G. Méndez-García LA, et al. Eur J Pediatr. 2021 Aug;180(8):2529-2537. doi: 10.1007/s00431-021-04104-0. Epub 2021 May 7. Eur J Pediatr. 2021. PMID: 33959818 - Exome sequencing analysis reveals variants in primary immunodeficiency genes in patients with very early onset inflammatory bowel disease.
Kelsen JR, Dawany N, Moran CJ, Petersen BS, Sarmady M, Sasson A, Pauly-Hubbard H, Martinez A, Maurer K, Soong J, Rappaport E, Franke A, Keller A, Winter HS, Mamula P, Piccoli D, Artis D, Sonnenberg GF, Daly M, Sullivan KE, Baldassano RN, Devoto M. Kelsen JR, et al. Gastroenterology. 2015 Nov;149(6):1415-24. doi: 10.1053/j.gastro.2015.07.006. Epub 2015 Jul 17. Gastroenterology. 2015. PMID: 26193622 Free PMC article. - miR-20a inhibits TCR-mediated signaling and cytokine production in human naïve CD4+ T cells.
Reddycherla AV, Meinert I, Reinhold A, Reinhold D, Schraven B, Simeoni L. Reddycherla AV, et al. PLoS One. 2015 Apr 17;10(4):e0125311. doi: 10.1371/journal.pone.0125311. eCollection 2015. PLoS One. 2015. PMID: 25884400 Free PMC article. - Association of Interleukin-10 -3575T>A and -1082A>G polymorphisms with non-Hodgkin lymphoma susceptibility: a comprehensive review and meta-analysis.
Zhang Y, Xia ZG, Zhu JH, Chen MB, Wang TM, Shen WX, He J. Zhang Y, et al. Mol Genet Genomics. 2015 Dec;290(6):2063-73. doi: 10.1007/s00438-015-1058-y. Epub 2015 May 15. Mol Genet Genomics. 2015. PMID: 25977148 Review. - Challenges and Prospects for Designer T and NK Cells in Glioblastoma Immunotherapy.
Arnesen VS, Gras Navarro A, Chekenya M. Arnesen VS, et al. Cancers (Basel). 2021 Oct 5;13(19):4986. doi: 10.3390/cancers13194986. Cancers (Basel). 2021. PMID: 34638471 Free PMC article. Review.
References
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. - PubMed
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–35. - PubMed
- Simonatto M, Natoli G. Functional genomics of the inflammatory response: Where are we now? Brief Funct Genomics. 2013 [Epub ahead of print] PMID: 23814131. - PubMed
- Sabat R, Grutz G, Warszawska K, et al. Biology of interleukin-10. Cytokine Growth Factor Rev. 2010;21:331–44. - PubMed
- Volk H, Asadullah K, Gallagher G, et al. IL-10 and its homologs: important immune mediators and emerging immunotherapeutic targets. Trends Immunol. 2001;22:414–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous