Signatures of mutational processes in human cancer - PubMed (original) (raw)
. 2013 Aug 22;500(7463):415-21.
doi: 10.1038/nature12477. Epub 2013 Aug 14.
Serena Nik-Zainal, David C Wedge, Samuel A J R Aparicio, Sam Behjati, Andrew V Biankin, Graham R Bignell, Niccolò Bolli, Ake Borg, Anne-Lise Børresen-Dale, Sandrine Boyault, Birgit Burkhardt, Adam P Butler, Carlos Caldas, Helen R Davies, Christine Desmedt, Roland Eils, Jórunn Erla Eyfjörd, John A Foekens, Mel Greaves, Fumie Hosoda, Barbara Hutter, Tomislav Ilicic, Sandrine Imbeaud, Marcin Imielinski, Natalie Jäger, David T W Jones, David Jones, Stian Knappskog, Marcel Kool, Sunil R Lakhani, Carlos López-Otín, Sancha Martin, Nikhil C Munshi, Hiromi Nakamura, Paul A Northcott, Marina Pajic, Elli Papaemmanuil, Angelo Paradiso, John V Pearson, Xose S Puente, Keiran Raine, Manasa Ramakrishna, Andrea L Richardson, Julia Richter, Philip Rosenstiel, Matthias Schlesner, Ton N Schumacher, Paul N Span, Jon W Teague, Yasushi Totoki, Andrew N J Tutt, Rafael Valdés-Mas, Marit M van Buuren, Laura van 't Veer, Anne Vincent-Salomon, Nicola Waddell, Lucy R Yates; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain; Jessica Zucman-Rossi, P Andrew Futreal, Ultan McDermott, Peter Lichter, Matthew Meyerson, Sean M Grimmond, Reiner Siebert, Elías Campo, Tatsuhiro Shibata, Stefan M Pfister, Peter J Campbell, Michael R Stratton
Collaborators, Affiliations
- PMID: 23945592
- PMCID: PMC3776390
- DOI: 10.1038/nature12477
Signatures of mutational processes in human cancer
Ludmil B Alexandrov et al. Nature. 2013.
Erratum in
- Nature. 2013 Oct 10;502(7470):258. Imielinsk, Marcin [corrected to Imielinski, Marcin]
Abstract
All cancers are caused by somatic mutations; however, understanding of the biological processes generating these mutations is limited. The catalogue of somatic mutations from a cancer genome bears the signatures of the mutational processes that have been operative. Here we analysed 4,938,362 mutations from 7,042 cancers and extracted more than 20 distinct mutational signatures. Some are present in many cancer types, notably a signature attributed to the APOBEC family of cytidine deaminases, whereas others are confined to a single cancer class. Certain signatures are associated with age of the patient at cancer diagnosis, known mutagenic exposures or defects in DNA maintenance, but many are of cryptic origin. In addition to these genome-wide mutational signatures, hypermutation localized to small genomic regions, 'kataegis', is found in many cancer types. The results reveal the diversity of mutational processes underlying the development of cancer, with potential implications for understanding of cancer aetiology, prevention and therapy.
Figures
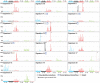
Figure 1. The prevalence of somatic mutations across human cancer types
Every dot represents a sample while the red horizontal lines are the median numbers of mutations in the respective cancer types. The vertical axis (log scaled) shows the number of mutations per megabase while the different cancer types are ordered on the horizontal axis based on their median numbers of somatic mutations. We would like to thank Gad Getz and colleagues for the design of this figure.
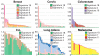
Figure 2. Validated mutational signatures found in human cancer
Each signature is displayed according to the 96 substitution classification defined by the substitution class and sequence context immediately 3′ and 5′ to the mutated base. The probability bars for the six types of substitutions are displayed in different colors. The mutation types are on the horizontal axes, while vertical axes depict the percentage of mutations attributed to a specific mutation type. All mutational signatures are displayed based on the trinucleotide frequency of the human genome. A higher resolution of each panel is found respectively in Supplementary Figures 2 to 23.
Figure 3. The presence of mutational signatures across human cancer types
Cancer types are ordered alphabetically as columns while mutational signatures are displayed as rows. “Other” indicates mutational signatures for which we were not able to perform validation or for which validation failed (Supplementary Figs 24 to 28). Prevalence in cancer samples indicates the percentage of samples from our dataset of 7,042 cancers in which the signature contributed significant number of somatic mutations. For most signatures, significant number of mutations in a sample is defined as more than 100 substitutions or more than 25% of all mutations in that sample.
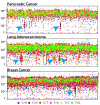
Figure 4. The contributions of mutational signatures to individual cancers of selected cancer types
Each bar represents a typical selected sample from the respective cancer type and the vertical axis denotes the number of mutations per megabase. Contributions across all cancer samples could be found in Supplementary Figures 29 to 58. Summary of the total contributions for all operative mutational processes in a cancer type could be found in Supplementary Figures 59 to 88. “Other” indicates mutational signatures for which we were not able to perform validation or for which validation failed (Supplementary Figs 24 to 28).
Figure 5. Mutational signatures with strong transcriptional strand bias
Mutations are shown according to the 192 mutation classification incorporating the substitution type, the sequence context immediately 5′ and 3′ to the mutated base and whether the mutated pyrimidine is on the transcribed or untranscribed strand. The mutation types are displayed on the horizontal axis, while the vertical axis depicts the percentage of mutations attributed to a specific mutation type. A higher resolution version of each panel is found respectively in Supplementary Figures 89 to 95.
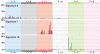
Figure 6. Kataegis in three cancers
Each of these “rainfall” plots represents an individual cancer sample in which each dot represents a single somatic mutation ordered on the horizontal axis according to its position in the human genome. The vertical axis denotes the genomic distance of each mutation from the previous mutation. Clusters of mutations in kataegis are arrowed.
Comment in
- Cancer genomics: Signature analysis suggests cancer origins.
Lokody I. Lokody I. Nat Rev Genet. 2013 Oct;14(10):677. doi: 10.1038/nrg3579. Epub 2013 Aug 28. Nat Rev Genet. 2013. PMID: 23982625 No abstract available. - Genetics: Signatures of mutational processes in cancer-a big step closer.
Hutchinson L. Hutchinson L. Nat Rev Clin Oncol. 2013 Oct;10(10):545. doi: 10.1038/nrclinonc.2013.162. Epub 2013 Sep 3. Nat Rev Clin Oncol. 2013. PMID: 23999215 No abstract available. - Genomics: Mutator catalogues.
Alderton GK. Alderton GK. Nat Rev Cancer. 2013 Oct;13(10):681. doi: 10.1038/nrc3608. Nat Rev Cancer. 2013. PMID: 24060860 No abstract available.
Similar articles
- Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer.
Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR. Nik-Zainal S, et al. Nat Genet. 2014 May;46(5):487-91. doi: 10.1038/ng.2955. Epub 2014 Apr 13. Nat Genet. 2014. PMID: 24728294 Free PMC article. - Learning mutational signatures and their multidimensional genomic properties with TensorSignatures.
Vöhringer H, Hoeck AV, Cuppen E, Gerstung M. Vöhringer H, et al. Nat Commun. 2021 Jun 15;12(1):3628. doi: 10.1038/s41467-021-23551-9. Nat Commun. 2021. PMID: 34131135 Free PMC article. - Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis.
Petljak M, Alexandrov LB, Brammeld JS, Price S, Wedge DC, Grossmann S, Dawson KJ, Ju YS, Iorio F, Tubio JMC, Koh CC, Georgakopoulos-Soares I, Rodríguez-Martín B, Otlu B, O'Meara S, Butler AP, Menzies A, Bhosle SG, Raine K, Jones DR, Teague JW, Beal K, Latimer C, O'Neill L, Zamora J, Anderson E, Patel N, Maddison M, Ng BL, Graham J, Garnett MJ, McDermott U, Nik-Zainal S, Campbell PJ, Stratton MR. Petljak M, et al. Cell. 2019 Mar 7;176(6):1282-1294.e20. doi: 10.1016/j.cell.2019.02.012. Cell. 2019. PMID: 30849372 Free PMC article. - Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair.
Phillips DH. Phillips DH. DNA Repair (Amst). 2018 Nov;71:6-11. doi: 10.1016/j.dnarep.2018.08.003. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30236628 Free PMC article. Review. - APOBEC-Induced Mutagenesis in Cancer.
Mertz TM, Collins CD, Dennis M, Coxon M, Roberts SA. Mertz TM, et al. Annu Rev Genet. 2022 Nov 30;56:229-252. doi: 10.1146/annurev-genet-072920-035840. Epub 2022 Aug 26. Annu Rev Genet. 2022. PMID: 36028227 Review.
Cited by
- Genomic characterization of small cell carcinomas of the uterine cervix.
Schultheis AM, de Bruijn I, Selenica P, Macedo GS, da Silva EM, Piscuoglio S, Jungbluth AA, Park KJ, Klimstra DS, Wardelmann E, Hartmann W, Gerharz CD, von Petersdorff M, Buettner R, Reis-Filho JS, Weigelt B. Schultheis AM, et al. Mol Oncol. 2022 Feb;16(4):833-845. doi: 10.1002/1878-0261.12962. Epub 2021 May 13. Mol Oncol. 2022. PMID: 33830625 Free PMC article. - Industrializing engineered autologous T cells as medicines for solid tumours.
Britten CM, Shalabi A, Hoos A. Britten CM, et al. Nat Rev Drug Discov. 2021 Jun;20(6):476-488. doi: 10.1038/s41573-021-00175-8. Epub 2021 Apr 8. Nat Rev Drug Discov. 2021. PMID: 33833444 Review. - Hypermutation takes the driver's seat.
Schlesner M, Eils R. Schlesner M, et al. Genome Med. 2015 Mar 28;7(1):31. doi: 10.1186/s13073-015-0159-x. eCollection 2015. Genome Med. 2015. PMID: 25821521 Free PMC article. - SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair.
Kanu N, Grönroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J, Maya-Mendoza A, Mistrík M, Rowan AJ, Patel H, Rabinowitz A, East P, Wilson G, Santos CR, McGranahan N, Gulati S, Gerlinger M, Birkbak NJ, Joshi T, Alexandrov LB, Stratton MR, Powles T, Matthews N, Bates PA, Stewart A, Szallasi Z, Larkin J, Bartek J, Swanton C. Kanu N, et al. Oncogene. 2015 Nov 12;34(46):5699-708. doi: 10.1038/onc.2015.24. Epub 2015 Mar 2. Oncogene. 2015. PMID: 25728682 Free PMC article. - Classification of Muscle-Invasive Bladder Cancer Based on Immunogenomic Profiling.
Zhou X, Qiu S, Nie L, Jin D, Jin K, Zheng X, Yang L, Wei Q. Zhou X, et al. Front Oncol. 2020 Aug 18;10:1429. doi: 10.3389/fonc.2020.01429. eCollection 2020. Front Oncol. 2020. PMID: 32974156 Free PMC article.
References
- Pena-Diaz J, et al. Noncanonical mismatch repair as a source of genomic instability in human cells. Molecular cell. 2012;47:669–680. - PubMed
Additional References (Online Methods)
Publication types
MeSH terms
Substances
Grants and funding
- 088340/WT_/Wellcome Trust/United Kingdom
- 093867/WT_/Wellcome Trust/United Kingdom
- T32 CA009216/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom