To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems - PubMed (original) (raw)
Review
To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems
Iwona Mruk et al. Nucleic Acids Res. 2014 Jan.
Abstract
One of the simplest classes of genes involved in programmed death is that containing the toxin-antitoxin (TA) systems of prokaryotes. These systems are composed of an intracellular toxin and an antitoxin that neutralizes its effect. These systems, now classified into five types, were initially discovered because some of them allow the stable maintenance of mobile genetic elements in a microbial population through postsegregational killing or the death of cells that have lost these systems. Here, we demonstrate parallels between some TA systems and restriction-modification systems (RM systems). RM systems are composed of a restriction enzyme (toxin) and a modification enzyme (antitoxin) and limit the genetic flux between lineages with different epigenetic identities, as defined by sequence-specific DNA methylation. The similarities between these systems include their postsegregational killing and their effects on global gene expression. Both require the finely regulated expression of a toxin and antitoxin. The antitoxin (modification enzyme) or linked protein may act as a transcriptional regulator. A regulatory antisense RNA recently identified in an RM system can be compared with those RNAs in TA systems. This review is intended to generalize the concept of TA systems in studies of stress responses, programmed death, genetic conflict and epigenetics.
Figures
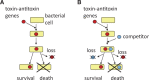
Figure 1.
Postsegregational killing or genetic addiction. (A) A TA system enters a bacterial cell and establishes itself in the bacterial lineage. Loss of the TA system from the cell triggers the death of its descendant cells. The TA system appears to be stably maintained in the population of viable cells. (B) Competitive advantage of TA systems. A cell maintains a TA system on a genetic element. A competitor genetic element enters the cell so that the two genetic elements are both present in the cell, although they are mutually exclusive in the long term. The loss of the competitor element (left) allows the survival of the descendant cell with the TA system, whereas the loss of the element with the TA system (right) leads to the death of the descendant cells, together with the competitor genetic element. The genetic element and the competitor genetic element can be two alleles of a single genomic locus.
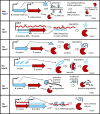
Figure 2.
TA systems and Type II RM systems. The toxin is a protein in all cases. The rightmost parts indicate the toxin action after the loss of the genes or an imbalance between the toxin and the antitoxin. Type II RMs have antitoxin modification enzymes that methylate the genomic DNA, protecting it from cleavage by a restriction enzyme (toxin); type I TA has an antisense RNA (the antitoxin) that pairs with the toxin mRNA (19,20); type II TA has a protein antitoxin that binds to the toxin (21,22); type III TA has an antitoxin RNA (not an antisense RNA) that binds to the toxin (23,24); type IV TA has a protein antitoxin that binds to the toxin target (25); and type V TA has a protein antitoxin that cleaves the toxin mRNA (26). A, antitoxin; T, toxin; P, promoter; Me, methyl group on a DNA base; SD, Shine–Dalgarno sequence. Toxins and their genes are shown in dark red; antitoxins and their genes are shown in light blue.
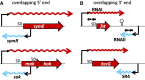
Figure 3.
Organization of type I TA systems. A region of the sense mRNA (rightward wavy line) encoding a protein toxin overlaps the coding region of an antitoxin antisense RNA (leftward wavy line) at their 5′ ends (A) or 3′ ends (B). A bar bent rightward above the gene represents a promoter, and a bar bent leftward beneath the gene represents a reverse promoter. In the _fst_-RNAI–RNAII system, the direct repeats and stem–loop structure are indicated. The figures are not to scale. Modified from: symE–symR (49), hok–sok (106), _fst_-RNAI–RNAII (54) and _bsrG_–SR4 (60). SD, Shine–Dalgarno sequence.
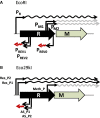
Figure 4.
Organization of RM systems with antisense RNA. The promoters (bent bars) and the mRNAs (rightward wavy lines) generated from them are shown above the restriction (R) and modification (M) genes, and the reverse promoters and the antisense RNAs are shown beneath them. A black wavy line represents a bicistronic mRNA, whereas a gray line represents the mRNA for a modification enzyme. A leftward wavy line indicates an antisense RNA. In (A), the transcription from PREV0 terminates at PM1M2. The termination sites for the antisense RNAs from PREV1/PREV2 and AS_P1/AS_P2 are unknown. The figures are not to scale.
Similar articles
- Structural Variations and Rearrangements in Bacterial Type II Toxin-Antitoxin Systems.
Nielsen MR, Brodersen DE. Nielsen MR, et al. Subcell Biochem. 2024;104:245-267. doi: 10.1007/978-3-031-58843-3_11. Subcell Biochem. 2024. PMID: 38963490 Review. - Homologous VapC Toxins Inhibit Translation and Cell Growth by Sequence-Specific Cleavage of tRNAfMet.
Walling LR, Butler JS. Walling LR, et al. J Bacteriol. 2018 Jan 10;200(3):e00582-17. doi: 10.1128/JB.00582-17. Print 2018 Feb 1. J Bacteriol. 2018. PMID: 29109187 Free PMC article. - Bacterial type I toxin-antitoxin systems.
Brantl S. Brantl S. RNA Biol. 2012 Dec;9(12):1488-90. doi: 10.4161/rna.23045. Epub 2012 Dec 1. RNA Biol. 2012. PMID: 23324552 - Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7.
Wilbaux M, Mine N, Guérout AM, Mazel D, Van Melderen L. Wilbaux M, et al. J Bacteriol. 2007 Apr;189(7):2712-9. doi: 10.1128/JB.01679-06. Epub 2007 Jan 26. J Bacteriol. 2007. PMID: 17259320 Free PMC article. - Why so narrow: Distribution of anti-sense regulated, type I toxin-antitoxin systems compared with type II and type III systems.
Coray DS, Wheeler NE, Heinemann JA, Gardner PP. Coray DS, et al. RNA Biol. 2017 Mar 4;14(3):275-280. doi: 10.1080/15476286.2016.1272747. Epub 2017 Jan 9. RNA Biol. 2017. PMID: 28067598 Free PMC article. Review.
Cited by
- The biocommunication method: On the road to an integrative biology.
Witzany G. Witzany G. Commun Integr Biol. 2016 Apr 8;9(2):e1164374. doi: 10.1080/19420889.2016.1164374. eCollection 2016 Mar-Apr. Commun Integr Biol. 2016. PMID: 27195071 Free PMC article. - Emerging Roles of Toxin-Antitoxin Modules in Bacterial Pathogenesis.
Kędzierska B, Hayes F. Kędzierska B, et al. Molecules. 2016 Jun 17;21(6):790. doi: 10.3390/molecules21060790. Molecules. 2016. PMID: 27322231 Free PMC article. Review. - Effects of Population Dynamics on Establishment of a Restriction-Modification System in a Bacterial Host.
Graovac S, Rodic A, Djordjevic M, Severinov K, Djordjevic M. Graovac S, et al. Molecules. 2019 Jan 7;24(1):198. doi: 10.3390/molecules24010198. Molecules. 2019. PMID: 30621083 Free PMC article. - Conserved DNA Methyltransferases: A Window into Fundamental Mechanisms of Epigenetic Regulation in Bacteria.
Oliveira PH, Fang G. Oliveira PH, et al. Trends Microbiol. 2021 Jan;29(1):28-40. doi: 10.1016/j.tim.2020.04.007. Epub 2020 May 13. Trends Microbiol. 2021. PMID: 32417228 Free PMC article. Review. - de novo synthesis of a bacterial toxin/antitoxin system.
Soo VW, Cheng HY, Kwan BW, Wood TK. Soo VW, et al. Sci Rep. 2014 May 6;4:4807. doi: 10.1038/srep04807. Sci Rep. 2014. PMID: 24797297 Free PMC article.
References
- Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics. 2004;272:227–234. - PubMed
- Kobayashi I. Genetic addiction-a principle in symbiosis of genes in a genome. In: Phillips G, Funnell B, editors. Plasmid Biology. Washington, DC: ASM Press; 2004. pp. 105–144.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases