DNA methylation markers in the postnatal developing rat brain - PubMed (original) (raw)
DNA methylation markers in the postnatal developing rat brain
Rebecca K Simmons et al. Brain Res. 2013.
Abstract
In spite of intense interest in how altered epigenetic processes including DNA methylation may contribute to psychiatric and neurodevelopmental disorders, there is a limited understanding of how methylation processes change during early postnatal brain development. The present study used in situ hybridization to assess mRNA expression for the three major DNA methyltranserases (DNMTs)--DNMT1, DNMT3a and DNMT3b--in the developing rat brain at seven developmental timepoints: postnatal days (P) 1, 4, 7, 10, 14, 21, and 75. We also assessed 5-methylcytosine levels (an indicator of global DNA methylation) in selected brain regions during the first three postnatal weeks. DNMT1, DNMT3a and DNMT3b mRNAs are widely expressed throughout the adult and postnatal developing rat brain. Overall, DNMT mRNA levels reached their highest point in the first week of life and gradually decreased over the first three postnatal weeks within the hippocampus, amygdala, striatum, cingulate and lateral septum. Global DNA methylation levels did not follow this developmental pattern; methylation levels gradually increased over the first three postnatal weeks in the hippocampus, and remained stable in the developing amygdala and prefrontal cortex. Our results contribute to a growing understanding of how DNA methylation markers unfold in the developing brain, and highlight how these developmental processes may differ within distinct brain regions.
Keywords: Amygdala; DNA methylation; DNA methyltransferase; Hippocampus; In situ hybridization; Neurodevelopment.
© 2013 Elsevier B.V. All rights reserved.
Figures
Fig. 1
DNMT1, DNMT3a and DNMT3b mRNA expression in the adult rat brain. Columns show autoradiograms from representative tissue sections through the adult rat brain processed by in situ hybridization with antisense probes against rat DNMT1 (left), DNMT3a (middle) and DNMT3b (right) mRNA. X-ray films were exposed for 8 days for DNMT1, 3 days for DNMT3a, and 17 days for DNMT3b. Images show examples of coronal sections through the adult Sprague–Dawley rat brain, depicting regions such as the hippocampus, amygdala, caudate putamen, and hypothalamus. The diagrams in the far right column (adapted from Paxinos and Watson, 1997) indicate anatomical landmarks for each section. Abbreviations can be found in Table 1.
Fig. 2
DNMT1, DNMT3a and DNMT3b mRNA expression in the developing rat brain. Columns show autoradiograms from representative tissue sections through the developmental rat brain at postnatal day (P)1, P4, P7, P10, P14, and P21 that were processed by in situ hybridization with antisense probe against rat DNMT1 (top column), DNMT3a (middle column) and DNMT3b (bottom column) mRNA. X-ray films were exposed for 8 days for DNMT1, 3 days for DNMT3a, and 17 days for DNMT3b.
Fig. 3
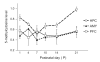
Developmental patterns of DNMT1, DNMT3a and DNMT3b mRNA expression in the early rodent postnatal period and adulthood. DNA methyltransferase (DNMT)1, DNMT3a, and DNMT3b mRNA expression levels were quantified in several regions of rat brains collected at 6 developmental timepoints – postnatal day (P)1, P4, P7, P10, P14, P21, and P75 (adulthood). (A) Developmental expression pattern of DNMT1, DNMT3a, and DNTM3b in subregions of the hippocampus (CA1–3 and dentate gyrus). (B) Developmental expression pattern of DNMT1, DNMT3a, and DNMT3b in subnuclei of the amygdala (lateral, basolateral, medial and central nucleus). (C) Developmental expression pattern of DNMT1 and DNMT3a in several forebrain regions, including cingulate (Cg) and lateral septum (LS). Due to low abundance, DNMT3b was unable to be quantified in these regions. (D) Developmental expression pattern of DNMT1 and DNMT3a in the striatum (caudate putamen (Cpu) and nucleus accumbens (Acb)). Due to low abundance, DNMT3b was unable to be quantified in these regions.
Fig. 4
Patterns of global DNA methylation in select brain regions of the developing rat brain. Global DNA methylation levels (approximated by measuring levels of 5-methylcytosine) were measured at several postnatal time periods – postnatal day (P)1, P4, P7, P10, P14 and P21 in tissue punches from the hippocampus, amygdala, and prefrontal cortex. Global DNA methylation levels appeared to increase, although this effect is most apparent in the developing hippocampus. Also, when comparing global DNA methylation levels across the three brain regions, levels were significantly higher in hippocampus compared to amygdala and prefrontal cortex.
Similar articles
- DNA methylation in the developing hippocampus and amygdala of anxiety-prone versus risk-taking rats.
Simmons RK, Howard JL, Simpson DN, Akil H, Clinton SM. Simmons RK, et al. Dev Neurosci. 2012;34(1):58-67. doi: 10.1159/000336641. Epub 2012 May 8. Dev Neurosci. 2012. PMID: 22572572 Free PMC article. - The loss of global DNA methylation due to decreased DNMT expression in the postnatal mouse ovaries may associate with infertility emerging during ovarian aging.
Uysal F, Ozturk S. Uysal F, et al. Histochem Cell Biol. 2020 Sep;154(3):301-314. doi: 10.1007/s00418-020-01890-w. Epub 2020 Jun 8. Histochem Cell Biol. 2020. PMID: 32514790 - Sex difference in the expression of DNA methyltransferase 3a in the rat amygdala during development.
Kolodkin MH, Auger AP. Kolodkin MH, et al. J Neuroendocrinol. 2011 Jul;23(7):577-83. doi: 10.1111/j.1365-2826.2011.02147.x. J Neuroendocrinol. 2011. PMID: 21518035 Free PMC article. - Role of Mammalian DNA Methyltransferases in Development.
Chen Z, Zhang Y. Chen Z, et al. Annu Rev Biochem. 2020 Jun 20;89:135-158. doi: 10.1146/annurev-biochem-103019-102815. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815535 Review. - Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma.
Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Hervouet E, et al. Clin Epigenetics. 2018 Feb 9;10:17. doi: 10.1186/s13148-018-0450-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449903 Free PMC article. Review.
Cited by
- DNA Methyltransferases in Depression: An Update.
Duan Z, Lu J. Duan Z, et al. Front Psychiatry. 2020 Sep 3;11:538683. doi: 10.3389/fpsyt.2020.538683. eCollection 2020. Front Psychiatry. 2020. PMID: 33101076 Free PMC article. Review. - Global DNA Methylation in the Chestnut Blight Fungus Cryphonectria parasitica and Genome-Wide Changes in DNA Methylation Accompanied with Sectorization.
So KK, Ko YH, Chun J, Bal J, Jeon J, Kim JM, Choi J, Lee YH, Huh JH, Kim DH. So KK, et al. Front Plant Sci. 2018 Feb 2;9:103. doi: 10.3389/fpls.2018.00103. eCollection 2018. Front Plant Sci. 2018. PMID: 29456549 Free PMC article. - Role of DNA methylation and the DNA methyltransferases in learning and memory.
Morris MJ, Monteggia LM. Morris MJ, et al. Dialogues Clin Neurosci. 2014 Sep;16(3):359-71. doi: 10.31887/DCNS.2014.16.3/mmorris. Dialogues Clin Neurosci. 2014. PMID: 25364286 Free PMC article. Review. - Maternal Style Selectively Shapes Amygdalar Development and Social Behavior in Rats Genetically Prone to High Anxiety.
Cohen JL, Glover ME, Pugh PC, Fant AD, Simmons RK, Akil H, Kerman IA, Clinton SM. Cohen JL, et al. Dev Neurosci. 2015;37(3):203-14. doi: 10.1159/000374108. Epub 2015 Mar 17. Dev Neurosci. 2015. PMID: 25791846 Free PMC article. - Nurture outpaces nature: fostering with an attentive mother alters social dominance in a mouse model of stress sensitivity.
Sur D, Agranyoni O, Kirby M, Cohen N, Bagaev A, Karandasheva K, Shmerkin E, Gorobets D, Savita BK, Avneri R, Divon MS, Lax E, Michaelevski I, Pinhasov A. Sur D, et al. Mol Psychiatry. 2023 Sep;28(9):3816-3828. doi: 10.1038/s41380-023-02273-y. Epub 2023 Oct 16. Mol Psychiatry. 2023. PMID: 37845494
References
- Agnish ND, Keller KA. The rationale for culling of rodent litters. Fundam. Appl. Toxicol.: Off. J. Soc. Toxicol. 1997;38:2–6. - PubMed
- Akbarian S. Epigenetics of schizophrenia. Curr. Top. Behav. Neurosci. 2010;4:611–628. - PubMed
- Beneyto M, Meador-Woodruff JH. Expression of transcripts encoding AMPA receptor subunits and associated postsynaptic proteins in the macaque brain. J. Comp. Neurol. 2004;468:530–554. - PubMed
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci.: MN. 2007;31:47–58. - PubMed
- Brown SE, Weaver IC, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci. Lett. 2008;440:49–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases