Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens - PubMed (original) (raw)
Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens
Patrick N Reardon et al. J Biol Chem. 2013.
Abstract
Several species of δ proteobacteria are capable of reducing insoluble metal oxides as well as other extracellular electron acceptors. These bacteria play a critical role in the cycling of minerals in subsurface environments, sediments, and groundwater. In some species of bacteria such as Geobacter sulfurreducens, the transport of electrons is proposed to be facilitated by filamentous fibers that are referred to as bacterial nanowires. These nanowires are polymeric assemblies of proteins belonging to the type IVa family of pilin proteins and are mainly comprised of one subunit protein, PilA. Here, we report the high resolution solution NMR structure of the PilA protein from G. sulfurreducens determined in detergent micelles. The protein is >85% α-helical and exhibits similar architecture to the N-terminal regions of other non-conductive type IVa pilins. The detergent micelle interacts with the first 21 amino acids of the protein, indicating that this region likely associates with the bacterial inner membrane prior to fiber formation. A model of the G. sulfurreducens pilus fiber is proposed based on docking of this structure into the fiber model of the type IVa pilin from Neisseria gonorrhoeae. This model provides insight into the organization of aromatic amino acids that are important for electrical conduction.
Keywords: Bacterial Metabolism; Bioenergetics; Electron Transport; Membrane Proteins; NMR; Pili; Structural Biology.
Figures
FIGURE 1.
Alignment of various major pilin subunit amino acid sequences. Species whose pilin subunit amino acid sequence length is >66 were truncated to 67 amino acids. Eight species capable of EET and two species that are not capable of EET were included in the alignment.
FIGURE 2.
Overview of the solution NMR structure of G. sulfurreducens PilA. A, overlay of the ensemble of 18 structures that did not contain NOE violations >0.5 Å or dihedral angle violations >5°. B, ribbon diagram of the selected conformer (see “Materials and Methods”) with the highly conserved core domain corresponding to amino acids 1–22 colored in orange and the rest of the protein colored blue. C, ribbon diagram of the selected conformer with the aromatic residues shown in blue space filling. The amino terminus and carboxyl terminus are indicated by N and C, respectively. D, 15N TROSY spectrum of GSu PilA with backbone amide assignments. Data were collected at 750 MHz on an Agilent VNMR spectrometer. E, overlay of the homology model of GSu PilA (38) and the experimentally determined GSu PilA structure. The arrows indicate where the degree of bend differs between the structures near residues 22 and 42. To emphasize the differences, the structures were aligned using residues 23–41.
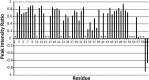
FIGURE 3.
Summary of H-N heteronuclear NOE data. Spectra with and without proton saturation (3s) were acquired. The plot shows the ratio of cross-peak intensities with and without proton saturation. The data were collected at 800 MHz. Asterisks indicate resonances that could not be clearly identified and quantitatively characterized.
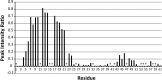
FIGURE 4.
Summary of paramagnetic relaxation by Gd-DTPA. The site-specific reduction in peak intensity upon addition of 4 m
m
Gd-DTPA to an 15N-labeled sample of GSu PilA is shown. Values near 1 indicate no loss of intensity and protection from Gd-DTPA. No correction was made for the small sample dilution (∼3% maximum) that occurred during the titration. The data were collected at 750 MHz. Asterisks indicate resonances that could not be clearly identified and quantitatively characterized.
FIGURE 5.
Model of a nanowire fiber based on the structure of G. sulfurreducens PilA. A, superimposition of PilA from G. sulfurreducens on to the homologous type IV pilin from N. gonorrhoeae (Protein Data Bank code 2HIL) (12). The amino terminus and carboxyl terminus are indicated by N and C, respectively. B, model of the bacterial nanowire, based on the pilus assembly of N. gonorrhoeae (Protein Data Bank code 2HIL) (12). Aromatic side chains are shown in space filling. A single cluster of aromatic side chains is shown in blue space filling, whereas all others are shown in gray. Ribbons of each subunit were colored individually. C, schematic diagram showing the progression of the aromatic clusters up the pilus structure. The aromatic band is colored blue, and the aromatic devoid band is colored yellow.
Similar articles
- Generation of High Current Densities in Geobacter sulfurreducens Lacking the Putative Gene for the PilB Pilus Assembly Motor.
Ueki T, Walker DJF, Nevin KP, Ward JE, Woodard TL, Nonnenmann SS, Lovley DR. Ueki T, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0087721. doi: 10.1128/Spectrum.00877-21. Epub 2021 Sep 29. Microbiol Spectr. 2021. PMID: 34585977 Free PMC article. - Bottom-Up Fabrication of Protein Nanowires via Controlled Self-Assembly of Recombinant Geobacter Pilins.
Cosert KM, Castro-Forero A, Steidl RJ, Worden RM, Reguera G. Cosert KM, et al. mBio. 2019 Dec 10;10(6):e02721-19. doi: 10.1128/mBio.02721-19. mBio. 2019. PMID: 31822587 Free PMC article. - Significance of a Posttranslational Modification of the PilA Protein of Geobacter sulfurreducens for Surface Attachment, Biofilm Formation, and Growth on Insoluble Extracellular Electron Acceptors.
Richter LV, Franks AE, Weis RM, Sandler SJ. Richter LV, et al. J Bacteriol. 2017 Mar 28;199(8):e00716-16. doi: 10.1128/JB.00716-16. Print 2017 Apr 15. J Bacteriol. 2017. PMID: 28138101 Free PMC article. - Biology and biotechnology of microbial pilus nanowires.
Clark MM, Reguera G. Clark MM, et al. J Ind Microbiol Biotechnol. 2020 Oct;47(9-10):897-907. doi: 10.1007/s10295-020-02312-5. Epub 2020 Oct 3. J Ind Microbiol Biotechnol. 2020. PMID: 33009965 Review. - Microbial nanowires for bioenergy applications.
Malvankar NS, Lovley DR. Malvankar NS, et al. Curr Opin Biotechnol. 2014 Jun;27:88-95. doi: 10.1016/j.copbio.2013.12.003. Epub 2013 Dec 31. Curr Opin Biotechnol. 2014. PMID: 24863901 Review.
Cited by
- The structures of two archaeal type IV pili illuminate evolutionary relationships.
Wang F, Baquero DP, Su Z, Beltran LC, Prangishvili D, Krupovic M, Egelman EH. Wang F, et al. Nat Commun. 2020 Jul 9;11(1):3424. doi: 10.1038/s41467-020-17268-4. Nat Commun. 2020. PMID: 32647180 Free PMC article. - The Pilin N-terminal Domain Maintains Neisseria gonorrhoeae Transformation Competence during Pilus Phase Variation.
Obergfell KP, Seifert HS. Obergfell KP, et al. PLoS Genet. 2016 May 23;12(5):e1006069. doi: 10.1371/journal.pgen.1006069. eCollection 2016 May. PLoS Genet. 2016. PMID: 27213957 Free PMC article. - Phylogenetic and structural diversity of aromatically dense pili from environmental metagenomes.
Bray MS, Wu J, Padilla CC, Stewart FJ, Fowle DA, Henny C, Simister RL, Thompson KJ, Crowe SA, Glass JB. Bray MS, et al. Environ Microbiol Rep. 2020 Feb;12(1):49-57. doi: 10.1111/1758-2229.12809. Epub 2019 Nov 21. Environ Microbiol Rep. 2020. PMID: 31701641 Free PMC article. - The major subunit of widespread competence pili exhibits a novel and conserved type IV pilin fold.
Sheppard D, Berry JL, Denise R, Rocha EPC, Matthews S, Pelicic V. Sheppard D, et al. J Biol Chem. 2020 May 8;295(19):6594-6604. doi: 10.1074/jbc.RA120.013316. Epub 2020 Apr 9. J Biol Chem. 2020. PMID: 32273343 Free PMC article. - Thermophiles; or, the Modern Prometheus: The Importance of Extreme Microorganisms for Understanding and Applying Extracellular Electron Transfer.
Lusk BG. Lusk BG. Front Microbiol. 2019 Apr 26;10:818. doi: 10.3389/fmicb.2019.00818. eCollection 2019. Front Microbiol. 2019. PMID: 31080440 Free PMC article.
References
- Lovley D. R., Holmes D. E., Nevin K. P. (2004) Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49, 219–286 - PubMed
- Lovley D. R., Phillips E. J. P., Gorby Y. A., Landa E. R. (1991) Microbial reduction of uranium. Nature 350, 413–416
- Myers C. R., Nealson K. H. (1988) Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321 - PubMed
- Lovley D. R., Coates J. D., Blunt-Harris E. L., Phillips E. J. P., Woodward J. C. (1996) Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources