Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer - PubMed (original) (raw)
Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer
Justin M Balko et al. Cancer Res. 2013.
Abstract
Basal-like breast cancer (BLBC) is an aggressive disease that lacks a clinically approved targeted therapy. Traditional chemotherapy is effective in BLBC, but it spares the cancer stem cell (CSC)-like population, which is likely to contribute to cancer recurrence after the initial treatment. Dual specificity phosphatase-4 (DUSP4) is a negative regulator of the mitogen-activated protein kinase (MAPK) pathway that is deficient in highly aggressive BLBCs treated with chemotherapy, leading to aberrant MAPK activation and resistance to taxane-induced apoptosis. Herein, we investigated how DUSP4 regulates the MAP-ERK kinase (MEK) and c-jun-NH2-kinase (JNK) pathways in modifying CSC-like behavior. DUSP4 loss increased mammosphere formation and the expression of the CSC-promoting cytokines interleukin (IL)-6 and IL-8. These effects were caused in part by loss of control of the MEK and JNK pathways and involved downstream activation of the ETS-1 and c-JUN transcription factors. Enforced expression of DUSP4 reduced the CD44(+)/CD24(-) population in multiple BLBC cell lines in a MEK-dependent manner, limiting tumor formation of claudin-low SUM159PT cells in mice. Our findings support the evaluation of MEK and JNK pathway inhibitors as therapeutic agents in BLBC to eliminate the CSC population.
©2013 AACR.
Conflict of interest statement
Conflicts of interest: N/A
Figures
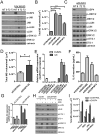
Figure 1. Low DUSP4 and high MEK activation is associated with tumors and cell lines with CSC-like features
A) Cancer Cell Line Encyclopedia data SNP copy number analysis for the DUSP4 locus across 964 cancer cell lines from various tissue sources (49). Red bars show the median log2 copy number for each tissue origin. B) Left panel; frequency histogram of TCGA (14) SNP copy number analysis for the DUSP4 locus for 443 breast cancers normalized to patient matched control tissue (log2 ratio). Right panel; DUSP4 log2 copy number ratio plotted by molecular subtype as determined by gene expression using the PAM50 centroids. P-value represents the ANOVA result. C) CD44:CD24 mRNA ratio for the ICBP50 panel of breast cancer cell lines, as determined by microarray data, plotted vs. the MEK activation score of Pratilas et al. (20). D) Unsupervised clustering of RNA from 15 mammosphere cultures vs. 11 primary tumor profiles, representing 16 patients including 10 patient pairs (7) using the genes defined in the MEK signature (20). E) Box and whisker plot of the data in (D), according to the total sum score of the MEK signature. P-value represents the result of a two-tailed t-test. F) CD44:CD24 mRNA ratio for the ICBP50 panel of breast cancer cell lines, as determined by microarray data plotted vs. DUSP4 mRNA expression. Points and text in black represent the correlation for all cell lines, and points and text in red represent the correlation of only the cell lines with a high MEK activation score (>median of all cell lines). G) Immunoblot analysis of a panel of breast cancer cell lines. TNBC: triple negative breast cancer; L: luminal, A: Basal A; B: Basal B.
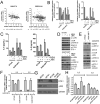
Figure 2. Loss of DUSP4 function upregulates IL-6 and IL-8 and enhances mammosphere growth
A) Immunoblot of MDA-231 cells 96 hrs after siCONTROL or siDUSP4 transfection in 10% FBS containing medium or after 24 hr of serum starvation in 0.1% FBS containing medium B) Mammosphere growth assay in MDA-231 cells, quantitated by GelCount software 6 days after siRNA transfection (5 days after plating to mammosphere culture conditions; ***p<0.001 for a two-tailed t-test). C) Immunoblot analysis performed on lysates from MDA-231 cells grown under non-adherent (mammosphere) conditions. D) Serum-free media was collected 72-96 hrs after siRNA transfection of MDA-231 cells and normalized to cell number. Conditioned medium was added to SUM159PT cells cultured in a mammosphere assay for 5 days and quantitated by GelCount software (*p=<0.05, two-tailed t-test E) qRT-PCR analysis of DUSP4 and IL6 mRNA expression in MDA-231 cells 72 hr after siRNA transfection. F) IL6 ELISA in serum-free medium conditioned by siRNA-transfected MDA-231 cells G) qRT-PCR analysis of IL6 mRNA expression in MDA-231 cells 96 hr after siRNA transfection and 4 hr after treatment with 1 μM selumetinib (MEKi) or 10 μM SP600125 (JNKi). H) Immunoblot analysis of MDA-231 cells after treatment for 24 hrs with 1 μM selumetinib (MEKi) or 10 μM SP600125 (JNKi). I. MDA-231 mammosphere formation quantitated by GelCount software 7 days after siRNA transfection. Where indicated, selumetinib (MEKi) or SP600125 (JNK1) or the combination was added to the mammosphere cultures. All bars represent the mean of 3 replicates ± SD.
Figure 3. DUSP4 regulates ETS-1 and cJUN and downstream IL-6 and IL-8 expression via MEK and JNK
A) Correlation of IL6 mRNA and IL8 mRNA with DUSP4 mRNA in 2 external microarray datasets of primary breast cancer. B) qRT-PCR analysis of BT549 and SUM159PT cells treated for 16 hr with 10 μM SP600125 (JNKi), 1 μM selumetinib (MEKi) or the combination. C) IL6 and IL8 ELISA analyses of conditioned media from BT549 and SUM159PT cells collected after 24-48 hr of treatment with the indicated inhibitors. D) Immunoblot analysis of lysates from cells shown in (B) and (C). E) Immunoblot analysis of BT549 and SUM159PT cells after 16 hr of adenovirus transduction with AdDUSP4 or AdGFP control. F) qRT-PCR analysis of cells from (E). G) Immunoblot analysis of SUM159PT harvested 96 hr after transfection with siCONTROL, siETS-1, siCJUN or the combination. H) qRT-PCR analysis of IL6 and IL8 mRNAs in cells from (G).
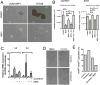
Figure 4. MEK inhibition decreases mammosphere formation in an IL-6/IL-8 dependent manner
A) Representative images of mammospheres derived from SUM159PT and BT549 cells grown in the presence of selumetinib or DMSO. B) Quantification of mammospheres derived from SUM159PT and BT549 cells, grown in the presence of DMSO, selumetinib (MEKi), or selumetinib plus either IL6, IL8, or both IL6 and IL8. P-value represents the result of an ANOVA with Tukey's post hoc test to compare individual treatment groups (***P<0.001). Bars represent mean of 3 replicates ± SD. C) qRT-PCR analysis of IL6 and IL8 mRNAs in MDA-231 xenografts treated for 3 days with docetaxel, selumetinib or the combination (16). D) MDA-231 xenografts treated for 4 weeks with docetaxel, selumetinib or the combination(16) were harvested; cells were mechanically and enzymatically dissociated and plated in a mammosphere assay as described in Methods. E) CD44/CD24 FACS analysis of cells dissociated from MDA-231 xenografts from (D).
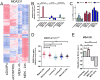
Figure 5. Gene expression patterns of loss of DUSP4 function correlate with basal-like and claudin-low subtypes
Microarray analysis was performed on RNA derived from MDA-231, BT549 and SUM159PT cells 96 hr after transfection with siCONTROL or siDUSP4 and after 4 hr or 24 hr of selumetinib treatment. A) Heatmap of significantly altered genes for MDA-231 cells (ANOVA FDR-adjusted p<0.01). B) DUSP4 loss gene expression score derived from MDA-231 cells transfected with siDUSP4 was calculated across all samples. C) Expression of selected chemotherapy-resistance genes which were altered in MDA-231 cells transfected with siDUSP4. D) DUSP4-loss gene expression score derived from MDA-231 cells transfected with siDUSP4 was calculated across 444 tumors in the TCGA breast data. Molecular subtype was determined using the PAM50 centroids (50) and the genefu package in R. E) Association of the claudin-low nine-cell line predictive genes with MDA-231 cells following siDUSP4 or selumetinib treatment (5).
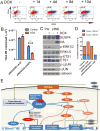
Figure 6. DUSP4 is a tumor suppressor that modulates the CD44+/CD24- stem-enriched population and tumor formation
SUM159PT cells transduced with DOX-inducible DUSP4-HA were generated. A) Flow cytometry analysis of CD44/CD24 expression in cells treated for 1-10 days with DOX (2 ng/mL). B) SUM159PT/pINDUCER-LACZ cells or SUM159PT/pINDUCER-DUSP4 pre-treated for 4 days ± DOX prior to plating in a mammosphere assay in the presence or absence of DOX as in the pretreatment. C) Immunoblot of parental SUM159PT/pINDUCER-LACZ cells or SUM159PT/pINDUCER-DUSP4 treated for 4 days with DOX. D) Tumor formation of cells from (C) 60 days after injection the mammary fatpad of athymic mice. Red bars indicate palpable tumors and blue bars indicate tumors identified on histological examination of the injection site. E) Schematic of proposed model based on the presented data: The tumor suppressor DUSP4 negatively regulates ERK and JNK. Upon loss of DUSP4, derepressed ERK and JNK activity stimulates ETS1 and cJUN-mediated transcription of IL6 and IL8, cytokines that expand the cancer stem-like cell population. De-repressed ERK transcription following DUSP4 loss also suppresses CD24 expression thus increasing the CD44HI CD24LO compartment, a marker of the CSC population.
Similar articles
- Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance.
Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, Sanders ME, Granja-Ingram NM, Smith JJ, Meszoely IM, Salter J, Dowsett M, Stemke-Hale K, González-Angulo AM, Mills GB, Pinto JA, Gómez HL, Arteaga CL. Balko JM, et al. Nat Med. 2012 Jul;18(7):1052-9. doi: 10.1038/nm.2795. Nat Med. 2012. PMID: 22683778 Free PMC article. - Statins affect ETS1-overexpressing triple-negative breast cancer cells by restoring DUSP4 deficiency.
Jung HH, Lee SH, Kim JY, Ahn JS, Park YH, Im YH. Jung HH, et al. Sci Rep. 2016 Sep 8;6:33035. doi: 10.1038/srep33035. Sci Rep. 2016. PMID: 27604655 Free PMC article. - Metalloprotease-dependent activation of EGFR modulates CD44+/CD24- populations in triple negative breast cancer cells through the MEK/ERK pathway.
Wise R, Zolkiewska A. Wise R, et al. Breast Cancer Res Treat. 2017 Nov;166(2):421-433. doi: 10.1007/s10549-017-4440-0. Epub 2017 Aug 8. Breast Cancer Res Treat. 2017. PMID: 28791489 Free PMC article. - Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling.
Caunt CJ, Keyse SM. Caunt CJ, et al. FEBS J. 2013 Jan;280(2):489-504. doi: 10.1111/j.1742-4658.2012.08716.x. Epub 2012 Aug 28. FEBS J. 2013. PMID: 22812510 Free PMC article. Review. - DUSP9, a Dual-Specificity Phosphatase with a Key Role in Cell Biology and Human Diseases.
Khoubai FZ, Grosset CF. Khoubai FZ, et al. Int J Mol Sci. 2021 Oct 26;22(21):11538. doi: 10.3390/ijms222111538. Int J Mol Sci. 2021. PMID: 34768967 Free PMC article. Review.
Cited by
- Single-cell transcriptional atlas of human breast cancers and model systems.
Altman JE, Olex AL, Zboril EK, Walker CJ, Boyd DC, Myrick RK, Hairr NS, Koblinski JE, Puchalapalli M, Hu B, Dozmorov MG, Chen XS, Chen Y, Perou CM, Lehmann BD, Visvader JE, Harrell JC. Altman JE, et al. Clin Transl Med. 2024 Oct;14(10):e70044. doi: 10.1002/ctm2.70044. Clin Transl Med. 2024. PMID: 39417215 Free PMC article. - Integrative transcriptome analysis reveals the molecular events underlying impaired T-cell responses in EGFR-mutant lung cancer.
Zhao Y, Tang G, Li J, Bian X, Zhou X, Feng J. Zhao Y, et al. Sci Rep. 2024 Aug 7;14(1):18366. doi: 10.1038/s41598-024-69020-3. Sci Rep. 2024. PMID: 39112565 Free PMC article. - From Tyrosine Kinases to Tyrosine Phosphatases: New Therapeutic Targets in Cancers and Beyond.
Zhou Y, Yao Z, Lin Y, Zhang H. Zhou Y, et al. Pharmaceutics. 2024 Jul 1;16(7):888. doi: 10.3390/pharmaceutics16070888. Pharmaceutics. 2024. PMID: 39065585 Free PMC article. Review. - Energy‑stress‑mediated activation of AMPK sensitizes MPS1 kinase inhibition in triple‑negative breast cancer.
Lim JS, Kim E, Song JS, Ahn S. Lim JS, et al. Oncol Rep. 2024 Aug;52(2):101. doi: 10.3892/or.2024.8760. Epub 2024 Jun 21. Oncol Rep. 2024. PMID: 38904203 Free PMC article. - From mechanism to therapy: the journey of CD24 in cancer.
Zhao K, Wu C, Li X, Niu M, Wu D, Cui X, Zhao H. Zhao K, et al. Front Immunol. 2024 May 31;15:1401528. doi: 10.3389/fimmu.2024.1401528. eCollection 2024. Front Immunol. 2024. PMID: 38881902 Free PMC article. Review.
References
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. - PubMed
- Blick T, Hugo H, Widodo E, Waltham M, Pinto C, Mani SA, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:235–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99/R00 CA142899/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- K99 CA142899/CA/NCI NIH HHS/United States
- P50 CA098131/CA/NCI NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- R01 CA143126/CA/NCI NIH HHS/United States
- P30 CA68465/CA/NCI NIH HHS/United States
- R01CA143126/CA/NCI NIH HHS/United States
- R00 CA142899/CA/NCI NIH HHS/United States
- P30 DK58404/DK/NIDDK NIH HHS/United States
- P50 CA98131/CA/NCI NIH HHS/United States
- P30 EY08126/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous