Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011 - PubMed (original) (raw)
Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011
Saskia L Smits et al. Emerg Infect Dis. 2013.
Abstract
To identify unknown human viruses, we analyzed serum and cerebrospinal fluid samples from patients with unexplained paraplegia from Malawi by using viral metagenomics. A novel cyclovirus species was identified and subsequently found in 15% and 10% of serum and cerebrospinal fluid samples, respectively. These data expand our knowledge of cyclovirus diversity and tropism.
Keywords: Malawi; Paraplegia; cerebrospinal fluid; cyclovirus; random amplification; serum; virus; viruses.
Figures
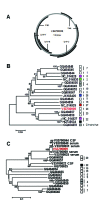
Figure
Cyclovirus genome organization and phylogenetic analysis of translated putative replication-associated protein (Rep) sequences. A) Size and predicted genome organization of human cyclovirus VS5700009, showing the 2 major open reading frames (ORFs) encoding Rep and the putative capsid protein (Cap, and other ORFs with a coding capacity >100 aa (ORFs 3–5). B) Phylogenetic tree of the translated Rep sequence of human cyclovirus VS5700009 and representative human and animal cycloviruses, generated by using MEGA5 with the neighbor-joining method with p-distance and 1,000 bootstrap replicates. A chimpanzee circovirus was used as an outgroup. Significant bootstrap values are shown. Cycloviruses in the same species are defined as having >85% aa identity in the Rep region and are labeled by vertical bars as described (11). The human (white), dragonfly (blue), bat (red), goat (pink), cattle (black), chicken (green), and chimpanzee (gray) cycloviruses are indicated, and VS5700009 is highlighted in red. Scale bar = 5% estimated phylogenetic divergence. C) Phylogenetic tree of the genomic area, corresponding to nt 392–733 in human cyclovirus VS5700009 and representative human and animal cycloviruses, generated by using MEGA5 with the neighbor-joining method with p-distance and 1,000 bootstrap replicates. Significant bootstrap values are shown. Cycloviruses isolated from the same species are labeled by vertical bars as described (11). Cycloviruses from humans (white), cattle (black), and chimpanzees (gray) are indicated, and VS5700009 is highlighted in red. Scale bar = 10% estimated phylogenetic divergence.
Similar articles
- Limited geographic distribution of the novel cyclovirus CyCV-VN.
Le VT, de Jong MD, Nguyen VK, Nguyen VT, Taylor W, Wertheim HF, van der Ende A, van der Hoek L, Canuti M, Crusat M, Sona S, Nguyen HU, Giri A, Nguyen TT, Ho DT, Farrar J, Bryant JE, Tran TH, Nguyen VV, van Doorn HR. Le VT, et al. Sci Rep. 2014 Feb 5;4:3967. doi: 10.1038/srep03967. Sci Rep. 2014. PMID: 24495921 Free PMC article. - Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections.
Tan le V, van Doorn HR, Nghia HD, Chau TT, Tu le TP, de Vries M, Canuti M, Deijs M, Jebbink MF, Baker S, Bryant JE, Tham NT, BKrong NT, Boni MF, Loi TQ, Phuong le T, Verhoeven JT, Crusat M, Jeeninga RE, Schultsz C, Chau NV, Hien TT, van der Hoek L, Farrar J, de Jong MD. Tan le V, et al. mBio. 2013 Jun 18;4(3):e00231-13. doi: 10.1128/mBio.00231-13. mBio. 2013. PMID: 23781068 Free PMC article. - High prevalence of cyclovirus Vietnam (CyCV-VN) in plasma samples from Madagascan healthy blood donors.
Sauvage V, Gomez J, Barray A, Vandenbogaert M, Boizeau L, Tagny CT, Rakoto O, Bizimana P, Guitteye H, Ciré BB, Soumana H, Tchomba JS, Caro V, Laperche S. Sauvage V, et al. Infect Genet Evol. 2018 Dec;66:9-12. doi: 10.1016/j.meegid.2018.09.002. Epub 2018 Sep 7. Infect Genet Evol. 2018. PMID: 30201501 - 2024 taxonomy update for the family Circoviridae.
Varsani A, Harrach B, Roumagnac P, Benkő M, Breitbart M, Delwart E, Franzo G, Kazlauskas D, Rosario K, Segalés J, Dunay E, Rukundo J, Goldberg TL, Fehér E, Kaszab E, Bányai K, Krupovic M. Varsani A, et al. Arch Virol. 2024 Aug 14;169(9):176. doi: 10.1007/s00705-024-06107-2. Arch Virol. 2024. PMID: 39143430 Review. - Recently discovered blood-borne viruses: are they hepatitis viruses or merely endosymbionts?
Mushahwar IK. Mushahwar IK. J Med Virol. 2000 Dec;62(4):399-404. doi: 10.1002/1096-9071(200012)62:4<399::aid-jmv1>3.0.co;2-u. J Med Virol. 2000. PMID: 11074465 Review. No abstract available.
Cited by
- T-cell reprogramming through targeted CD4-coreceptor and T-cell receptor expression on maturing thymocytes by latent Circoviridae family member porcine circovirus type 2 cell infections in the thymus.
Klausmann S, Sydler T, Summerfield A, Lewis FI, Weilenmann R, Sidler X, Brugnera E. Klausmann S, et al. Emerg Microbes Infect. 2015 Mar;4(3):e15. doi: 10.1038/emi.2015.15. Epub 2015 Mar 11. Emerg Microbes Infect. 2015. PMID: 26038767 Free PMC article. - Nontraumatic Myelopathy in Malawi: A Prospective Study in an Area with High HIV Prevalence.
Zijlstra EE, van Hellemond JJ, Moes AD, de Boer C, Boeschoten SA, van Blijswijk CEM, van der Vuurst de Vries RM, Bailey PAB, Kampondeni S, van Lieshout L, Smits SL, Katchanov J, Mkandawire NM, Rothe C. Zijlstra EE, et al. Am J Trop Med Hyg. 2020 Feb;102(2):451-457. doi: 10.4269/ajtmh.19-0209. Am J Trop Med Hyg. 2020. PMID: 31837130 Free PMC article. - Novel Cyclovirus Species in Dogs with Hemorrhagic Gastroenteritis.
Gainor K, Malik YS, Ghosh S. Gainor K, et al. Viruses. 2021 Oct 26;13(11):2155. doi: 10.3390/v13112155. Viruses. 2021. PMID: 34834961 Free PMC article. - Diversity of CRESS DNA Viruses in Squamates Recapitulates Hosts Dietary and Environmental Sources of Exposure.
Capozza P, Lanave G, Diakoudi G, Pellegrini F, Cardone R, Vasinioti VI, Decaro N, Elia G, Catella C, Alberti A, Bányai K, Mendoza-Roldan JA, Otranto D, Buonavoglia C, Martella V. Capozza P, et al. Microbiol Spectr. 2022 Jun 29;10(3):e0078022. doi: 10.1128/spectrum.00780-22. Epub 2022 May 26. Microbiol Spectr. 2022. PMID: 35616383 Free PMC article. - Plasma virome of cattle from forest region revealed diverse small circular ssDNA viral genomes.
Wang H, Li S, Mahmood A, Yang S, Wang X, Shen Q, Shan T, Deng X, Li J, Hua X, Cui L, Delwart E, Zhang W. Wang H, et al. Virol J. 2018 Jan 15;15(1):11. doi: 10.1186/s12985-018-0923-9. Virol J. 2018. PMID: 29334978 Free PMC article.
References
- Wallace ID, Cosnett JE. Unexplained spastic paraplegia. S Afr Med J. 1983;63:689–91 . - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources