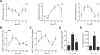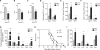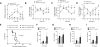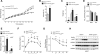Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes - PubMed (original) (raw)
Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes
Khoa D Nguyen et al. Science. 2013.
Abstract
Circadian clocks have evolved to regulate physiologic and behavioral rhythms in anticipation of changes in the environment. Although the molecular clock is present in innate immune cells, its role in monocyte homeostasis remains unknown. Here, we report that Ly6C(hi) inflammatory monocytes exhibit diurnal variation, which controls their trafficking to sites of inflammation. This cyclic pattern of trafficking confers protection against Listeria monocytogenes and is regulated by the repressive activity of the circadian gene Bmal1. Accordingly, myeloid cell-specific deletion of Bmal1 induces expression of monocyte-attracting chemokines and disrupts rhythmic cycling of Ly6C(hi) monocytes, predisposing mice to development of pathologies associated with acute and chronic inflammation. These findings have unveiled a critical role for BMAL1 in controlling the diurnal rhythms in Ly6C(hi) monocyte numbers.
Figures
Fig. 1. Diurnal variation of Ly6Chi monocytes
(A) Quantitative RT-PCR analysis of clock controlled genes (Arntl, Nr1d1, and Dbp) in blood monocytes during a 12 hour light-dark cycle (n = 3-4 samples per time point). (B and C) Ly6Chi monocyte numbers in blood (B) and spleen (C) during a 12 hour light-dark cycle (n=5 mice per time point). (D and E) Recruitment of Ly6Chi monocytes to inflamed peritoneum. Ly6Chi monocyte number (D), and concentration of IL1β (E) were quantified in the peritoneal fluid 2 hours after elicitation with thioglycollate (n = 5 mice per time point). Pooled data (A) or representatives (B to E) of two independent experiments are shown as mean ± SEM. Two-tailed Student's t-tests (A) and one-way ANOVA (B-E) are used for statistical analyses (comparisons were made between the acrophase and other time points). *P<0.05; **P<0.01; ***P<0.001.
Fig. 2. Diurnal variation in the pathogenicity of Listeria monocytogenes
(A to C) Mice kept under a 12 hour light-dark cycle were intraperitoneally inoculated with 1×106 L. monocytogenes at ZT0 and ZT8, and colony forming units (CFUs) recovered from the peritoneal cavity (A), spleen (B), and liver (C) were quantified 2 dpi (n = 14-15 mice per time point). (D to F) Numbers of iNOS+CD11c+ and TNFα+CD11c+ cells in peritoneal cavity (D), spleen (E), and liver (F) of mice 2 dpi with 1×106 L. monocytogenes at ZT0 and ZT8 (n = 15 mice per time point). (G) Concentration of chemokines and cytokines in peritoneal fluid 2 dpi with 1×106 L. monocytogenes at ZT0 and ZT8 (n = 15 mice per time point). (H) Survival curves of mice after infection with 1×107 L. monocytogenes at ZT0 and ZT8 (n = 25 mice per time point). (I) Serum concentration of chemokines and cytokines 2 dpi with 1×107 L. monocytogenes at ZT0 and ZT8 (n = 10 mice per time point). Pooled data from two or three independent experiments are presented as mean ± SEM. Statistical significance (*P<0.05; **P < 0.01; ***P < 0.001) was assessed using two-tailed Student's _t_-test (A to G, and I), and log-rank test (H).
Fig. 3. BMAL1 regulates rhythmic oscillations of Ly6Chi monocytes in a cell autonomous manner
(A) Quantitative RT-PCR kinetic analysis of Nr1d1 mRNA in blood monocytes isolated from _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice kept on a 12 hour light-dark cycle (n = 3-4 samples per genotype and time point). (B to D) Ly6Chi monocyte numbers in blood (B), spleen (C), and bone marrow (D) of _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice kept under a 12 hour light-dark cycle at various ZTs. (n = 5 mice per genotype and time point). (E) Survival curves of _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice after infection with 1×106 L. monocytogenes at ZT0 and ZT8 (n = mice 10-11 per genotype and time point). (F to I) Serum concentrations of IL1β (F), IL6 (G), IFNγ (H), and CCL2 (I) in _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice 2 dpi with 1×106 L. monocytogenes at ZT0 and ZT8 (n = mice 4-6 per genotype and time point). Pooled data (A and E) and representative (B to D) of two to three independent experiments are shown as mean ± SEM and analyzed using two-tailed Student's _t_-tests (A to D, and F to I), and log-rank test (E). *P<0.05; **P<0.01; ***P<0.001.
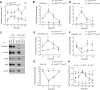
Fig. 4. BMAL1 recruits PRC2 to repress expression of Ccl2
(A) Quantitative RT-PCR analysis of Ccl2 expression in blood monocytes of _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice kept on a 12 hour day-ligh cycle (n = 3-4 samples per genotype and time point). (B) ChIP analysis of BMAL1 binding to the Ccl2 promoter (n = 4 samples per genotype and time point). (C) Coimmunoprecipitation of BMAL1 and with members of PRC2. Nuclear lysates from serum shocked BMDMs were immunopercipiated with BMAL1 antibody, and immunoblotted for BMAL1, CLOCK, EZH2, EED, and SUZ12. (D and G) ChIP analysis for the recruitment of EZH2 (D) and Pol II (G) to the proximal promoter of the Ccl2 gene (n = 4 samples per genotype and time point). (E and F) ChIP analysis for H3K27Me3 (E) and H3K4Me3 (F) at the proximal promoter of Ccl2 gene (n = 4 samples per genotype and time point). (H) Ly6Chi monocyte numbers in the blood of wild type or _Ccr2_−/− mice during a 12 hour light-dark cycle. Wild type mice were intraperitoneally injected with PBS (Veh) or CCL2 (20 μgkg−1) 24 hours prior to quantification of Ly6Chi monocytes (n = 4-5 mice per genotype/treatment and time point). Pooled data (A to G) from two independent experiments are shown as mean ± S.E.M and analyzed using two-tailed Student's _t_-tests (A and B; D to G) and two-way ANOVA (H). *P<0.05; **P<0.01; ***P<0.001 represent comparison between _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre or between wild type treated Veh vs. CCL2 or _Ccr2_−/− at each time point.
Fig. 5. Myeloid cell-specific deletion of BMAL1 exacerbates metabolic disease
(A to D) Body weight (A), adiposity (B), tissue weights (C) and oxygen consumption (D) in _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice kept on a 12 hour light-dark cycle fed high fat diet for 19 weeks (n = 4-5 mice per genotype). (E) Total and Ly6Chi macrophage content in eWAT of _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice fed high fat diet (n = 5 mice per genotype). (F and G) Glucose (F) and insulin tolerance (G) tests of _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre mice fed high fat diet (n = 5-8 mice per genotype). (H) Immunoblots of total and phosphorylated AKT (pAKT) in eWAT of obese _Arntl_LoxP/LoxP and _Arntl_LoxP/LoxP_Lyz2_Cre administered intraportal insulin. Representative data of two to four independent experiments (A to C, E to H) are shown as mean ± S.E.M and analyzed using two-tailed Student's _t_-tests. *P<0.05; **P<0.01; ***P<0.001.
Comment in
- Immunology. Some monocytes got rhythm.
Druzd D, Scheiermann C. Druzd D, et al. Science. 2013 Sep 27;341(6153):1462-4. doi: 10.1126/science.1244445. Science. 2013. PMID: 24072913 No abstract available.
Similar articles
- Myeloid Bmal1 deletion increases monocyte recruitment and worsens atherosclerosis.
Huo M, Huang Y, Qu D, Zhang H, Wong WT, Chawla A, Huang Y, Tian XY. Huo M, et al. FASEB J. 2017 Mar;31(3):1097-1106. doi: 10.1096/fj.201601030R. Epub 2016 Dec 7. FASEB J. 2017. PMID: 27927724 Free PMC article. - Clock gene Bmal1 controls diurnal rhythms in expression and activity of intestinal carboxylesterase 1.
Chen X, Yu F, Guo X, Su C, Li SS, Wu B. Chen X, et al. J Pharm Pharmacol. 2021 Mar 1;73(1):52-59. doi: 10.1093/jpp/rgaa035. J Pharm Pharmacol. 2021. PMID: 33791812 - Immunology. Some monocytes got rhythm.
Druzd D, Scheiermann C. Druzd D, et al. Science. 2013 Sep 27;341(6153):1462-4. doi: 10.1126/science.1244445. Science. 2013. PMID: 24072913 No abstract available. - Monocyte-mediated immune defense against murine Listeria monocytogenes infection.
Serbina NV, Shi C, Pamer EG. Serbina NV, et al. Adv Immunol. 2012;113:119-34. doi: 10.1016/B978-0-12-394590-7.00003-8. Adv Immunol. 2012. PMID: 22244581 Free PMC article. Review. - Circadian clocks: from stem cells to tissue homeostasis and regeneration.
Dierickx P, Van Laake LW, Geijsen N. Dierickx P, et al. EMBO Rep. 2018 Jan;19(1):18-28. doi: 10.15252/embr.201745130. Epub 2017 Dec 19. EMBO Rep. 2018. PMID: 29258993 Free PMC article. Review.
Cited by
- Role for Retinoic Acid-Related Orphan Receptor Alpha (RORα) Expressing Macrophages in Diet-Induced Obesity.
Hams E, Roberts J, Bermingham R, Hogan AE, O'Shea D, O'Neill L, Fallon PG. Hams E, et al. Front Immunol. 2020 Aug 27;11:1966. doi: 10.3389/fimmu.2020.01966. eCollection 2020. Front Immunol. 2020. PMID: 32973801 Free PMC article. - Regulation of emergency granulopoiesis during infection.
Paudel S, Ghimire L, Jin L, Jeansonne D, Jeyaseelan S. Paudel S, et al. Front Immunol. 2022 Sep 5;13:961601. doi: 10.3389/fimmu.2022.961601. eCollection 2022. Front Immunol. 2022. PMID: 36148240 Free PMC article. Review. - Timing is everything: impact of development, ageing and circadian rhythm on macrophage functions in urinary tract infections.
Wang AS, Steers NJ, Parab AR, Gachon F, Sweet MJ, Mysorekar IU. Wang AS, et al. Mucosal Immunol. 2022 Jun;15(6):1114-1126. doi: 10.1038/s41385-022-00558-z. Epub 2022 Aug 29. Mucosal Immunol. 2022. PMID: 36038769 Review. - IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system.
Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, Finck R, Carmi Y, Zunder ER, Fantl WJ, Bendall SC, Engleman EG, Nolan GP. Spitzer MH, et al. Science. 2015 Jul 10;349(6244):1259425. doi: 10.1126/science.1259425. Science. 2015. PMID: 26160952 Free PMC article. - Functions for Retinoic Acid-Related Orphan Receptor Alpha (RORα) in the Activation of Macrophages During Lipopolysaccharide-Induced Septic Shock.
Hams E, Roberts J, Bermingham R, Fallon PG. Hams E, et al. Front Immunol. 2021 Mar 9;12:647329. doi: 10.3389/fimmu.2021.647329. eCollection 2021. Front Immunol. 2021. PMID: 33767712 Free PMC article.
References
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011 Feb 2;13:125. - PubMed
- Saini C, Suter DM, Liani A, Gos P, Schibler U. The mammalian circadian timing system: synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol. 2011;76:39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP1AR064158/AR/NIAMS NIH HHS/United States
- T32AI007334/AI/NIAID NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- HL076746/HL/NHLBI NIH HHS/United States
- R01 HL076746/HL/NHLBI NIH HHS/United States
- P01AI063302/AI/NIAID NIH HHS/United States
- DP1 AR064158/AR/NIAMS NIH HHS/United States
- R01 DK094641/DK/NIDDK NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- DK094641/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases