Accurate SAXS profile computation and its assessment by contrast variation experiments - PubMed (original) (raw)
Accurate SAXS profile computation and its assessment by contrast variation experiments
Dina Schneidman-Duhovny et al. Biophys J. 2013.
Abstract
A major challenge in structural biology is to characterize structures of proteins and their assemblies in solution. At low resolution, such a characterization may be achieved by small angle x-ray scattering (SAXS). Because SAXS analyses often require comparing profiles calculated from many atomic models against those determined by experiment, rapid and accurate profile computation from molecular structures is needed. We developed fast open-source x-ray scattering (FoXS) for profile computation. To match the experimental profile within the experimental noise, FoXS explicitly computes all interatomic distances and implicitly models the first hydration layer of the molecule. For assessing the accuracy of the modeled hydration layer, we performed contrast variation experiments for glucose isomerase and lysozyme, and found that FoXS can accurately represent density changes of this layer. The hydration layer model was also compared with a SAXS profile calculated for the explicit water molecules in the high-resolution structures of glucose isomerase and lysozyme. We tested FoXS on eleven protein, one DNA, and two RNA structures, revealing superior accuracy and speed versus CRYSOL, AquaSAXS, the Zernike polynomials-based method, and Fast-SAXS-pro. In addition, we demonstrated a significant correlation of the SAXS score with the accuracy of a structural model. Moreover, FoXS utility for analyzing heterogeneous samples was demonstrated for intrinsically flexible XLF-XRCC4 filaments and Ligase III-DNA complex. FoXS is extensively used as a standalone web server as a component of integrative structure determination by programs IMP, Chimera, and BILBOMD, as well as in other applications that require rapidly and accurately calculated SAXS profiles.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Fit and residual plots for FoXS and CRYSOL: experimental data (dark gray), FoXS (red), and CRYSOL (green). The fit plots are for q(Å−1) (x axis) versus log-intensity (y axis). The residual plots are for q(Å−1) (x axis) versus experimental intensity divided by computed intensity (y axis). The cases are ordered by the number of atoms as in Tables 1 and 2.
Figure 2
Comparison of FoXS and CRYSOL adjustable parameters and running times. (A) CRYSOL _r_0 versus _c_1. (B) CRYSOL δρ versus FOXS _c_2. (C) Running times of FoXS (red) versus CRYSOL (green) and CRYSOL L50 (dark green, order of harmonics = 50).
Figure 3
Cross correlation of _χ_-scores with model accuracy. (A) Superoxide dismutase, (B) abscisic acid receptor PYR1, (C) ubiquitin-like modifier-activating enzyme ATG7 C-terminal domain, (D) DNA double-strand break repair protein MRE11, and (E) glucose isomerase. For each of the five cases, a set of models is shown, followed by the _χ_-scores versus model accuracy for FoXS, FoXS′, and CRYSOL.
Figure 4
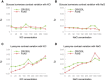
Salt concentration versus hydration layer density (δρ) for FoXS (red) and CRYSOL (green). (A and B) Glucose isomerase with KCl and NaCl, respectively. (C and D) Lysozyme with KCl and NaCl, respectively.
Figure 5
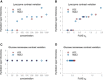
The thickness of hydration layer based on crystal waters versus concentration for (A) lysozyme and (C) glucose isomerase. The thickness of hydration layer based on crystal waters versus hydration layer density (FoXS _c_2) for (B) lysozyme and (D) glucose isomerase. (Blue) NaCl; (red) KCl.
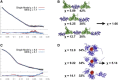
Figure 6
Finding the minimal ensemble consistent with a SAXS profile. Ensemble fit (blue) of the SAXS data (black) versus single conformation fit (red) for (A) XLF-XRCC4 filaments and (C) ligase III-DNA complex. (B and D) Conformations and their weights in the minimal ensemble.
Similar articles
- FoXS: a web server for rapid computation and fitting of SAXS profiles.
Schneidman-Duhovny D, Hammel M, Sali A. Schneidman-Duhovny D, et al. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W540-4. doi: 10.1093/nar/gkq461. Epub 2010 May 27. Nucleic Acids Res. 2010. PMID: 20507903 Free PMC article. - Pepsi-SAXS: an adaptive method for rapid and accurate computation of small-angle X-ray scattering profiles.
Grudinin S, Garkavenko M, Kazennov A. Grudinin S, et al. Acta Crystallogr D Struct Biol. 2017 May 1;73(Pt 5):449-464. doi: 10.1107/S2059798317005745. Epub 2017 Apr 27. Acta Crystallogr D Struct Biol. 2017. PMID: 28471369 - FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles.
Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Schneidman-Duhovny D, et al. Nucleic Acids Res. 2016 Jul 8;44(W1):W424-9. doi: 10.1093/nar/gkw389. Epub 2016 May 5. Nucleic Acids Res. 2016. PMID: 27151198 Free PMC article. - Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.
Ekimoto T, Ikeguchi M. Ekimoto T, et al. Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review. - A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins.
Kikhney AG, Svergun DI. Kikhney AG, et al. FEBS Lett. 2015 Sep 14;589(19 Pt A):2570-7. doi: 10.1016/j.febslet.2015.08.027. Epub 2015 Aug 29. FEBS Lett. 2015. PMID: 26320411 Review.
Cited by
- Unfolding bovine α-lactalbumin with T-jump: Characterizing disordered intermediates via time-resolved x-ray solution scattering and molecular dynamics simulations.
Hsu DJ, Leshchev D, Kosheleva I, Kohlstedt KL, Chen LX. Hsu DJ, et al. J Chem Phys. 2021 Mar 14;154(10):105101. doi: 10.1063/5.0039194. J Chem Phys. 2021. PMID: 33722011 Free PMC article. - Unusual Cytochrome _c_552 from Thioalkalivibrio paradoxus: Solution NMR Structure and Interaction with Thiocyanate Dehydrogenase.
Britikov VV, Bocharov EV, Britikova EV, Dergousova NI, Kulikova OG, Solovieva AY, Shipkov NS, Varfolomeeva LA, Tikhonova TV, Timofeev VI, Shtykova EV, Altukhov DA, Usanov SA, Arseniev AS, Rakitina TV, Popov VO. Britikov VV, et al. Int J Mol Sci. 2022 Sep 1;23(17):9969. doi: 10.3390/ijms23179969. Int J Mol Sci. 2022. PMID: 36077365 Free PMC article. - Computational Methods to Investigate Intrinsically Disordered Proteins and their Complexes.
Liu ZH, Tsanai M, Zhang O, Forman-Kay J, Head-Gordon T. Liu ZH, et al. ArXiv [Preprint]. 2024 Sep 3:arXiv:2409.02240v1. ArXiv. 2024. PMID: 39279844 Free PMC article. Preprint. - A Mechanically Weak Extracellular Membrane-Adjacent Domain Induces Dimerization of Protocadherin-15.
De-la-Torre P, Choudhary D, Araya-Secchi R, Narui Y, Sotomayor M. De-la-Torre P, et al. Biophys J. 2018 Dec 18;115(12):2368-2385. doi: 10.1016/j.bpj.2018.11.010. Epub 2018 Nov 16. Biophys J. 2018. PMID: 30527337 Free PMC article. - SSEThread: Integrative threading of the DNA-PKcs sequence based on data from chemical cross-linking and hydrogen deuterium exchange.
Saltzberg DJ, Hepburn M, Pilla KB, Schriemer DC, Lees-Miller SP, Blundell TL, Sali A. Saltzberg DJ, et al. Prog Biophys Mol Biol. 2019 Oct;147:92-102. doi: 10.1016/j.pbiomolbio.2019.09.003. Epub 2019 Sep 27. Prog Biophys Mol Biol. 2019. PMID: 31570166 Free PMC article.
References
- Putnam C.D., Hammel M., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083960/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- P01 CA092584/CA/NCI NIH HHS/United States
- PN2 EY016525/EY/NEI NIH HHS/United States
- R01GM105404/GM/NIGMS NIH HHS/United States
- R01 GM105404/GM/NIGMS NIH HHS/United States