The shared genomic architecture of human nucleolar organizer regions - PubMed (original) (raw)
The shared genomic architecture of human nucleolar organizer regions
Ioanna Floutsakou et al. Genome Res. 2013 Dec.
Abstract
The short arms of the five acrocentric human chromosomes harbor sequences that direct the assembly and function of the nucleolus, one of the key functional domains of the nucleus, yet they are absent from the current human genome assembly. Here we describe the genomic architecture of these human nucleolar organizers. Sequences distal and proximal to ribosomal gene arrays are conserved among the acrocentric chromosomes, suggesting they are sites of frequent recombination. Although previously believed to be heterochromatic, characterization of these two flanking regions reveals that they share a complex genomic architecture similar to other euchromatic regions of the genome, but they have distinct genomic characteristics. Proximal sequences are almost entirely segmentally duplicated, similar to the regions bordering centromeres. In contrast, the distal sequence is predominantly unique to the acrocentric short arms and is dominated by a very large inverted repeat. We show that the distal element is localized to the periphery of the nucleolus, where it appears to anchor the ribosomal gene repeats. This, combined with its complex chromatin structure and transcriptional activity, suggests that this region is involved in nucleolar organization. Our results provide a platform for investigating the role of NORs in nucleolar formation and function, and open the door for determining the role of these regions in the well-known empirical association of nucleoli with pathology.
Figures
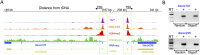
Figure 1.
Human rDNA flanking regions. (A) Schematic human acrocentric chromosome showing telomeric (blue) and centromeric (yellow) regions, and the NOR (black line), expanded below into rDNA, PJ (orange), and DJ (green) regions. Not to scale. (B) DJ and PJ localize distally and proximally to rDNA, respectively, on all acrocentric chromosomes. FISH was performed on normal human metaphase spreads with DJ BAC (red) and rDNA (green) probes (left panels), and PJ BAC (green) and rDNA (red) probes (right panel). Chromosomes are DAPI-stained. (C) DNA combing of HeLa cell nucleolar DNA shows DJ (red) is physically linked to 18S rDNA (green). Three representative images are shown below the hybridization scheme.
Figure 2.
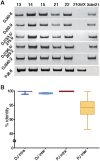
DJ and PJ acrocentric chromosome conservation. (A) PCR performed at increasing distances (left) into the DJ from mouse somatic cell hybrids carrying a single human acrocentric chromosome (indicated above). The right-hand lanes show PCR performed on the reciprocal products (Xder21 and 21derX) of a chr21 translocation that originates in the rDNA, confirming the DJ is located distally to the rDNA. Bottom panel is the same, but uses primers to the single unique PJ region. (B) Average intrachromosomal and interchromosomal DJ and PJ sequence identities from pairwise comparisons of representative BAC and cosmid clones are plotted.
Figure 3.
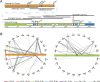
DJ and PJ sequence characterization. (A) Major genomic features of the PJ (orange) and DJ (green) contigs. BACs used to construct the contigs are shown as black lines, with BAC names and chromosomal origins indicated (chr17 annotation of AC011841 is incorrect). Satellites are shown in blue, the ACRO138 repeats in red, the large DJ inverted repeat as white arrows, and the rDNA array between the PJ and DJ in gray. (B) Segmental duplication analysis. Lines show segmental duplications from PJ (orange) and DJ (green), indicating the location of the duplicate on the human chromosomes (arranged around the flanking regions). Positions of centromeres (yellow) and telomeres (blue) are indicated. Segmental duplicate length is indicated by line color, as defined below.
Figure 4.
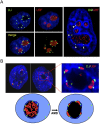
The DJ forms a perinucleolar anchor for rDNA repeats. (A) 3D immuno-FISH reveals that DJ sequences lie in perinucleolar heterochromatin in HT1080 cells. Nucleoli are visualized with UBF antibodies (red) and DJ with BAC CT476834 (green). The nucleus is DAPI-stained. The extended focus images (left) are stills from Supplemental Videos 1 and 2, while the image on the right shows a single focal plane. (B) Inhibition of rDNA transcription with AMD results in formation of nucleolar CAPs juxtaposed with DJ sequences in perinucleolar heterochromatin. Staining as in A. Two representative cells are shown, one with an enlargement. Cartoon models the transition between active and withdrawn rDNA upon AMD treatment. rDNA (red) retreats from the nucleolus (black) to the DJ (green) that is embedded in perinucleolar heterochromatin (dark blue).
Figure 5.
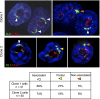
Ectopic DJ arrays target perinucleolar heterochromatin. Positioning of DJ BAC arrays. 3D FISH was performed on AMD-treated cells from clones 1 and 2 with rDNA (red) and DJ BAC CT476834 (green) probes. The large green hybridization signals identified by arrowheads indicate the ectopic DJ array. Endogenous DJ signals are also visible. Classification of ectopic DJ arrays as nucleolar associated, partially associated, or nonassociated is indicated by white, yellow, and orange arrowheads, respectively, and is quantified below.
Figure 6.
Chromatin landscape of the DJ. (A) ChIP-seq signals of different chromatin features (right) in H1-hESC cells, normalized to tags per million mapped reads are shown below a schematic of the DJ, including inverted repeats. Asterisks indicate enrichment sites. (Bottom) Control signal is shown in gray. (B) Chromatin states derived from the multivariate HMM analysis for seven different cell types (right). Each colored bar represents a specific chromatin state, as annotated below left. (C) Nucleolar H3K4me3 ChIP-PCR and nucleolar FAIRE-PCR using HT1080 cells validate the presence of H3K4me3 and FAIRE in the DJ. DJ positions of the primers used are shown to the right, and red boxes correspond to peaks of H3K4me3 from A. Genomic DNA (gDNA), input and negative controls (-ve and IgG) are shown.
Figure 7.
DJ transcript profiling. (A) ChIP-seq reveals chromatin features consistent with transcription originating from promoters at 187 kb and 238 kb (boxed) in the DJ. Top four tracks are an enlargement of selected chromatin features from Figure 6A. Bottom two tracks show RNA-seq and cDNA mapping results. Exons are indicated by blocks. These identify spliced transcripts (disnor187 and disnor238) similar to cDNA clones AK026938 and BX647690. (B) RT-PCR using primers to detect disnor187 and disnor238 transcripts in HT1080 cells. Random and oligo(dT)-primed RT-PCR products of the expected sizes for spliced transcripts were produced.
Similar articles
- The p-Arms of Human Acrocentric Chromosomes Play by a Different Set of Rules.
McStay B. McStay B. Annu Rev Genomics Hum Genet. 2023 Aug 25;24:63-83. doi: 10.1146/annurev-genom-101122-081642. Epub 2023 Feb 28. Annu Rev Genomics Hum Genet. 2023. PMID: 36854315 Review. - NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA.
van Sluis M, van Vuuren C, Mangan H, McStay B. van Sluis M, et al. Proc Natl Acad Sci U S A. 2020 May 12;117(19):10368-10377. doi: 10.1073/pnas.2001812117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332163 Free PMC article. - Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli.
Mangan H, Gailín MÓ, McStay B. Mangan H, et al. FEBS J. 2017 Dec;284(23):3977-3985. doi: 10.1111/febs.14108. Epub 2017 Jun 2. FEBS J. 2017. PMID: 28500793 Review. - Nucleolar organizer regions: genomic 'dark matter' requiring illumination.
McStay B. McStay B. Genes Dev. 2016 Jul 15;30(14):1598-610. doi: 10.1101/gad.283838.116. Genes Dev. 2016. PMID: 27474438 Free PMC article. Review. - Actively transcribed rDNA and distal junction (DJ) sequence are involved in association of NORs with nucleoli.
Liskovykh M, Petrov NS, Noskov VN, Masumoto H, Earnshaw WC, Schlessinger D, Shabalina SA, Larionov V, Kouprina N. Liskovykh M, et al. Cell Mol Life Sci. 2023 Apr 12;80(5):121. doi: 10.1007/s00018-023-04770-3. Cell Mol Life Sci. 2023. PMID: 37043028 Free PMC article.
Cited by
- Ribosomal DNA Instability as a Potential Cause of Karyotype Evolution.
Li D, Gandhi D, Kumon T, Yamashita YM. Li D, et al. Mol Biol Evol. 2022 Nov 3;39(11):msac221. doi: 10.1093/molbev/msac221. Mol Biol Evol. 2022. PMID: 36223491 Free PMC article. - A cohesin/HUSH- and LINC-dependent pathway controls ribosomal DNA double-strand break repair.
Marnef A, Finoux AL, Arnould C, Guillou E, Daburon V, Rocher V, Mangeat T, Mangeot PE, Ricci EP, Legube G. Marnef A, et al. Genes Dev. 2019 Sep 1;33(17-18):1175-1190. doi: 10.1101/gad.324012.119. Epub 2019 Aug 8. Genes Dev. 2019. PMID: 31395742 Free PMC article. - A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription.
Herdman C, Mars JC, Stefanovsky VY, Tremblay MG, Sabourin-Felix M, Lindsay H, Robinson MD, Moss T. Herdman C, et al. PLoS Genet. 2017 Jul 17;13(7):e1006899. doi: 10.1371/journal.pgen.1006899. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28715449 Free PMC article. - Determinants of mammalian nucleolar architecture.
Farley KI, Surovtseva Y, Merkel J, Baserga SJ. Farley KI, et al. Chromosoma. 2015 Sep;124(3):323-31. doi: 10.1007/s00412-015-0507-z. Epub 2015 Feb 12. Chromosoma. 2015. PMID: 25670395 Free PMC article. Review. - Choreographing the Double Strand Break Response: Ubiquitin and SUMO Control of Nuclear Architecture.
Harding SM, Greenberg RA. Harding SM, et al. Front Genet. 2016 Jun 7;7:103. doi: 10.3389/fgene.2016.00103. eCollection 2016. Front Genet. 2016. PMID: 27375678 Free PMC article. Review.
References
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 - PubMed
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M 2005. Nucleolar proteome dynamics. Nature 433: 77–83 - PubMed
- Bailey JA, Eichler EE 2006. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564 - PubMed
- Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D 1994. Alignment and sensitive detection of DNA by a moving interface. Science 265: 2096–2098 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials