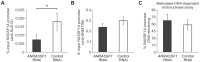The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation - PubMed (original) (raw)
The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation
Felipe C Beckedorff et al. PLoS Genet. 2013.
Abstract
The down-regulation of the tumor-suppressor gene RASSF1A has been shown to increase cell proliferation in several tumors. RASSF1A expression is regulated through epigenetic events involving the polycomb repressive complex 2 (PRC2); however, the molecular mechanisms modulating the recruitment of this epigenetic modifier to the RASSF1 locus remain largely unknown. Here, we identify and characterize ANRASSF1, an endogenous unspliced long noncoding RNA (lncRNA) that is transcribed from the opposite strand on the RASSF1 gene locus in several cell lines and tissues and binds PRC2. ANRASSF1 is transcribed through RNA polymerase II and is 5'-capped and polyadenylated; it exhibits nuclear localization and has a shorter half-life compared with other lncRNAs that bind PRC2. ANRASSF1 endogenous expression is higher in breast and prostate tumor cell lines compared with non-tumor, and an opposite pattern is observed for RASSF1A. ANRASSF1 ectopic overexpression reduces RASSF1A abundance and increases the proliferation of HeLa cells, whereas ANRASSF1 silencing causes the opposite effects. These changes in ANRASSF1 levels do not affect the RASSF1C isoform abundance. ANRASSF1 overexpression causes a marked increase in both PRC2 occupancy and histone H3K27me3 repressive marks, specifically at the RASSF1A promoter region. No effect of ANRASSF1 overexpression was detected on PRC2 occupancy and histone H3K27me3 at the promoter regions of RASSF1C and the four other neighboring genes, including two well-characterized tumor suppressor genes. Additionally, we demonstrated that ANRASSF1 forms an RNA/DNA hybrid and recruits PRC2 to the RASSF1A promoter. Together, these results demonstrate a novel mechanism of epigenetic repression of the RASSF1A tumor suppressor gene involving antisense unspliced lncRNA, in which ANRASSF1 selectively represses the expression of the RASSF1 isoform overlapping the antisense transcript in a location-specific manner. In a broader perspective, our findings suggest that other non-characterized unspliced intronic lncRNAs transcribed in the human genome might contribute to a location-specific epigenetic modulation of genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
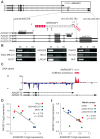
Figure 1. Antisense noncoding RNA ANRASSF1 is expressed within the RASSF1 genomic locus and inversely correlated with RASSF1 expression.
(A) Schematic representation of the entire RASSF1 locus presented at the top. Protein-coding RASSF1 gene isoforms (RefSeq-annotated) are shown in black; ANRASSF1 evidence from an assembly of public ESTs is shown in light gray; the portions extended through 5′-end RACE and primer-walking or 3′-end RACE are shown in red, with a poly(A) segment detected through sequencing. The arrowheads define the orientation of the sequences. (B) ANRASSF1 was detected in several human cell lines using end-point PCR in the antisense (AS), and not the sense (S), direction relative to the protein-coding gene. Strand-specific primers were used for the reverse-transcription (RT) (short half-arrow numbers 2 and 3 on ANRASSF1 in panel A). RT reaction in the absence of primer was used as a negative control (Ctl). The short half-arrows numbered 1 and 4 indicate the primers used for the end-point PCR. (C) Strand-specific RNA-seq from poly(A)+ RNA of LNCaP cells. The abundance of the reads mapping within the RASSF1 genomic locus is displayed in the diagram, and the color corresponds to the DNA strand of the transcripts. The red horizontal bar represents the consensus sequence from the assembly of RNA-seq reads mapping to the positive strand in this locus, generated using the Cufflinks tool. The gray bar represents the full-length ANRASSF1, obtained by RACE-PCR and Sanger sequencing. The arrowheads define the orientation of the sequences. (D) Expression of RASSF1 and ANRASSF1 measured with Affymetrix microarrays in HeLa, MDA-MB-231 and MCF-7 cells from GSE5823 . (E) The expression of RASSF1 and ANRASSF1 measured with Affymetrix microarrays in Jurkat cells under mitotic stress induced through treatment with phorbol ester and ionomycin for 30 and 60 min from GSE11118 .
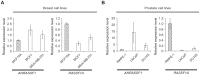
Figure 2. Inverse correlation between ANRASSF1 and RASSF1A expression in non-tumor and tumor cell lines.
Expression of ANRASSF1 and of RASSF1A in (A) breast and (B) prostate cell lines. Tumor cell lines (white bars) and non-tumor immortalized cell lines (hatched bars) were tested. These data show the means ± SD from two or three independent biological replicates for each cell line. The expression values in tumors are shown compared with the expression in the non-tumor cell line. These data are calculated relative to HPRT1 expression.
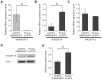
Figure 3. Overexpression of ANRASSF1 decreases the protein-coding RASSF1A isoform.
(A) Expression of ANRASSF1 in HeLa cells transfected with the _ANRASSF1_-containing vector (pCEP4 ANRASSF1) compared with ANRASSF1 expression in control HeLa cells transfected with an empty vector (empty pCEP4). Relative expression detected using RT-qPCR; these data are plotted relative to α-tubulin. These data show the means ± SD from three independent replicate transfection experiments. *p<0.01 relative to control. (B) Relative expression of RASSF1A isoform in _ANRASSF1_-transfected and control cells, as described in (A). These data show the means ± SD from three independent transfection experiments. *p<0.01 relative to control. (C) Relative expression of the RASSF1C isoform in _ANRASSF1_-transfected and control cells, as described in (A). These data show the means ± SD from three independent transfection experiments. (D) Western blot analysis using an antibody against RASSF1A protein in the lysates obtained from control HeLa cells (empty pCEP4) or _ANRASSF1_-transfected cells (pCEP4 ANRASSF1). An antibody against actin was used as control. (E) Densitometric analyses of the RASSF1A western blot signals from three replicate assays. Relative protein levels of RASSF1A normalized to actin. These data show the means ± SD from three independent transfection experiments. *p<0.02.
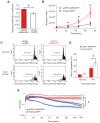
Figure 4. Overexpression of ANRASSF1 increases cell growth and decreases UVC- and staurosporine-mediated cell death.
(A) Cell proliferation coefficients of _ANRASSF1_-transfected cells (pCEP4 ANRASSF1) relative to control cells (empty pCEP4), as measured using MTS assay. These data show the means ± SD from three independent experiments. *_t_-test p<0.03 relative to control. (B) Effect of ANRASSF1 overexpression on the cell population growth, as measured by counting the number of cells over time in culture and calculating the population doubling time. These data show the means ± SD from two independent experiments in triplicate. * _t_-test p<0.05. (C) DNA content histograms showing the effect of ANRASSF1 overexpression on the cell cycle at 48 h after exposure to UVC irradiation (40 J/m2). The cells were labeled with propidium iodide. Light gray indicates the sub-G1 population. (D) Percent of sub-G1 population from experiments identical to (C). These data show the means ± SD from three independent replicate experiments. *_t_-test p<0.05 relative to control. (E) Effect of ANRASSF1 overexpression on cell survival in the presence of the cytotoxic drug staurosporine (100 nM). The changes in impedance were measured using the xCELLigence system to continuously monitor cell attachment to the culture plates. Three independent biological replicates for either condition (pCEP4 ANRASSF1 or empty pCEP4) are shown, each representing the means ± SD of two or three technical replicates; the red lines show cells overexpressing ANRASSF1, and the blue lines show control cells carrying empty pCEP4) vector. *_t_-test p<0.001, for the cell indices at 170 h.
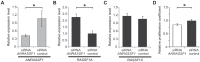
Figure 5. ANRASSF1 knockdown increases the RASSF1A isoform expression and decreases cell proliferation.
(A) Expression levels of ANRASSF1 in cells treated with siRNA ANRASSF1 relative to the expression of ANRASSF1 in control cells treated with scrambled siRNA (siRNA control). The relative expression was detected using RT-qPCR normalized to α-tubulin. These data show the means ± SD from three independent experiments. *p<0.01 relative to control. (B) The relative expression levels of RASSF1A in siRNA ANRASSF1 cells and siRNA control cells, as described in (A). These data show the means ± SD from three independent experiments. *p<0.01 relative to the control. (C) The relative expression levels of RASSF1C in siRNA ANRASSF1 cells and siRNA control cells, as described in (A). (D) Cell proliferation coefficients of siRNA ANRASSF1 cells relative to the control cells (siRNA control). These data show the means ± SD from three independent experiments. *p< 0.02 relative to control.
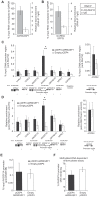
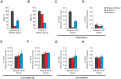
Figure 6. ANRASSF1 interacts with PRC2 and affects its occupancy at the RASSF1A promoter.
(A) Endogenous ANRASSF1 levels bound to PRC2 were measured in HeLa cells through RNA IP with anti-SUZ12 relative to the input. A control IP with non-specific IgG was performed in parallel. As a negative control, GAPDH mRNA, which was not expected to bind to PRC2, was used. The percent input in the IP fractions was shown as the ANRASSF1/GAPDH ratio. These data show the means ± SD from three independent experiments. (B) lincRNA S_FPQ_ is a positive control that binds to PRC2; RNA IP was assayed as in (A). These data show the means ± SD from three independent experiments. (C) ChIP assay using an anti-SUZ12 antibody in HeLa cells overexpressing ANRASSF1 (pCEP4 ANRASSF1, black bars) or control cells (empty pCEP4, white bars). The promoter regions of the RASSF1A and RASSF1C isoforms and two other genes on either side of the RASSF1 locus on chr 3 were investigated (the promoters are indicated with vertical lines in the scheme shown at the bottom of the figure). Control GAPDH was included as a gene not expected to be regulated through SUZ12. Control HOXA9 is a gene regulated through SUZ12 and encoded on chr 7. The amount of DNA in anti-SUZ12 samples at each promoter region detected through qPCR analysis was calculated in relation to the input. These data show the means ± SD from three independent experiments. *_t_-test p<0.02 relative to control at the RASSF1A locus. No significant changes were detected at other loci. (D) ChIP analysis using an anti-H3K27me3 antibody in an assay similar to that described in (C), except that the enrichment was calculated relative to anti-H3 ChIP. These data show the means ± SD from three independent experiments. *_t_-test p<0.02 relative to control at the RASSF1A locus. No significant changes were detected at the other loci. (E) ChIP analysis using an anti-DNMT3B antibody in an assay similar to that described in (C). These data show the means ± SD from three independent experiments. No significant change was observed. (F) DNA methylation at the RASSF1A promoter region was detected through qPCR with a methylation-dependent McrBC endonuclease assay in the _ANRASSF1_-overexpressing or control cells. The percentage of DNA remaining was calculated after comparing the amount of DNA amplified through qPCR following treatment with McrBC endonuclease with that following no-endonuclease treatment. These data show the means ± SD from three independent experiments. No significant change was observed.
Figure 7. ANRASSF1 mediates recruitment of SUZ12 to the RASSF1A promoter.
(A) RNase assay for detection of ANRASSF1 using RT-qPCR in permeabilized HeLa cells treated with RNase inhibitor (black bar), RNase H (red bar) or RNase A (blue bar). RNA% for each of the two RNase treatments was calculated relative to the corresponding values for the RNase inhibitor. These data show the means ± SD from three independent experiments. (B) As a control, alpha-tubulin mRNA was measured using RT-qPCR in parallel under the same conditions as described in (A). These data show the means ± SD from three independent experiments. (C) RNase-ChIP assay with anti-SUZ12 antibody in permeabilized HeLa cells treated with either RNase inhibitor (black bar), RNase H (red bar) or RNase A (blue bar). The amount of DNA at the RASSF1A promoter region detected through qPCR in anti-SUZ12 samples was calculated in relation to the input. These data show the means ± SD from two independent experiments that were performed in triplicate. (D) The amount of DNA at the RASSF1C promoter region was measured under the same conditions as described in (C). (E–F) The amount of DNA at the RASSF1A and RASSF1C promoter regions was measured under the same conditions as in (C–D), except that an anti-DNMT3B antibody was used. (G–H) The amount of DNA at the RASSF1A and RASSF1C promoter regions was measured under the same conditions as in (C–D), except that an anti-RNA Pol II antibody was used.
Figure 8. PRC2 is specifically directed by ANRASSF1 lncRNA to the RASSF1A promoter.
(A) ChIP assay using an anti-SUZ12 antibody in HeLa cells transfected with an RNAi oligonucleotide antisense to ANRASSF1 or with a control scrambled oligonucleotide. The amount of DNA at the RASSF1A promoter region detected using qPCR in the anti-SUZ12 samples was calculated in relation to the input. These data show the means ± SD from two independent experiments performed in triplicate. *p<0.03 relative to control. (B) ChIP assay using an anti-DNMT3B antibody in an assay similar to (A). (C) DNA methylation at the RASSF1A promoter region was detected through qPCR in a methylation-dependent McrBC endonuclease assay in cells transfected with an RNAi oligonucleotide antisense to ANRASSF1 or with a control scrambled oligonucleotide. The percent remaining DNA was calculated by comparing the amount of DNA at the promoter of RASSF1A amplified through qPCR following treatment with McrBC endonuclease against that amplified in the no-endonuclease treatment. These data show the means ± SD from three independent experiments. No significant change was observed.
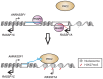
Figure 9. Proposed model for ANRASSF1 function at the RASSF1 genomic locus.
We postulate that lncRNA ANRASSF1 (blue line) interacts with genomic DNA at the transcription site, forming an RNA/DNA hybrid, and recruits the chromatin-modifying PRC2 complex to the protein-coding RASSF1A gene promoter region. The recruitment of the PCR2 complex results in the selective modification of the histone H3K27 pattern of methylation (red circles) at the RASSF1A promoter, leading to a specific reduction in RASSF1A transcriptional activity with no effect on the RASSF1C transcription.
Similar articles
- The long non-coding RNA ANRASSF1 in the regulation of alternative protein-coding transcripts RASSF1A and RASSF1C in human breast cancer cells: implications to epigenetic therapy.
Calanca N, Paschoal AP, Munhoz ÉP, Galindo LT, Barbosa BM, Caldeira JRF, Oliveira RA, Cavalli LR, Rogatto SR, Rainho CA. Calanca N, et al. Epigenetics. 2019 Aug;14(8):741-750. doi: 10.1080/15592294.2019.1615355. Epub 2019 May 27. Epigenetics. 2019. PMID: 31062660 Free PMC article. - Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma.
Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG. Byun DS, et al. Cancer Res. 2001 Oct 1;61(19):7034-8. Cancer Res. 2001. PMID: 11585730 - Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma.
Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG. Lee MG, et al. Cancer Res. 2001 Sep 15;61(18):6688-92. Cancer Res. 2001. PMID: 11559536 - Polycomb Gene Silencing Mechanisms: PRC2 Chromatin Targeting, H3K27me3 'Readout', and Phase Separation-Based Compaction.
Guo Y, Zhao S, Wang GG. Guo Y, et al. Trends Genet. 2021 Jun;37(6):547-565. doi: 10.1016/j.tig.2020.12.006. Epub 2021 Jan 22. Trends Genet. 2021. PMID: 33494958 Free PMC article. Review. - The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2.
Davidovich C, Cech TR. Davidovich C, et al. RNA. 2015 Dec;21(12):2007-22. doi: 10.1261/rna.053918.115. RNA. 2015. PMID: 26574518 Free PMC article. Review.
Cited by
- MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures.
Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. Mondal T, et al. Nat Commun. 2015 Jul 24;6:7743. doi: 10.1038/ncomms8743. Nat Commun. 2015. PMID: 26205790 Free PMC article. - Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment.
Pathania AS, Challagundla KB. Pathania AS, et al. Mol Ther Nucleic Acids. 2020 Oct 4;23:1371-1383. doi: 10.1016/j.omtn.2020.09.039. eCollection 2021 Mar 5. Mol Ther Nucleic Acids. 2020. PMID: 33738133 Free PMC article. Review. - Transcriptional gene silencing in humans.
Weinberg MS, Morris KV. Weinberg MS, et al. Nucleic Acids Res. 2016 Aug 19;44(14):6505-17. doi: 10.1093/nar/gkw139. Epub 2016 Apr 7. Nucleic Acids Res. 2016. PMID: 27060137 Free PMC article. - HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines.
Yunusov D, Anderson L, DaSilva LF, Wysocka J, Ezashi T, Roberts RM, Verjovski-Almeida S. Yunusov D, et al. Sci Rep. 2016 Sep 8;6:32753. doi: 10.1038/srep32753. Sci Rep. 2016. PMID: 27605307 Free PMC article. - Sex-specific variation in R-loop formation in Drosophila melanogaster.
Stanek TJ, Cao W, Mehra RM, Ellison CE. Stanek TJ, et al. PLoS Genet. 2022 Jun 10;18(6):e1010268. doi: 10.1371/journal.pgen.1010268. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35687614 Free PMC article.
References
- Donninger H, Vos MD, Clark GJ (2007) The RASSF1A tumor suppressor. J Cell Sci 120: 3163–3172. - PubMed
- Dammann R, Schagdarsurengin U, Strunnikova M, Rastetter M, Seidel C, et al. (2003) Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol Histopathol 18: 665–677. - PubMed
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, et al. (2000) Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25: 315–319. - PubMed
- Agathanggelou A, Cooper WN, Latif F (2005) Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res 65: 3497–3508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (SVA and EMR) and from Instituto Nacional de Ciência e Tecnologia em Oncogenômica (SVA and EMR), by fellowships from FAPESP (FCB, RCS, HIN, DTS, ACA) and from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (MSA) and by investigator fellowship awards (CNPq) (SVA, CFMM and EMR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources