An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules - PubMed (original) (raw)
. 2013 Aug 29;154(5):1151-1161.
doi: 10.1016/j.cell.2013.08.003.
Nicole E Bodycombe 1, Jaime H Cheah 1, Edmund V Price 1, Ke Liu 1, Giannina I Schaefer 1, Richard Y Ebright 1, Michelle L Stewart 1, Daisuke Ito 1, Stephanie Wang 1, Abigail L Bracha 1, Ted Liefeld 1, Mathias Wawer 1, Joshua C Gilbert 1, Andrew J Wilson 2, Nicolas Stransky 1, Gregory V Kryukov 1, Vlado Dancik 1, Jordi Barretina 1, Levi A Garraway 1, C Suk-Yee Hon 1, Benito Munoz 1, Joshua A Bittker 1, Brent R Stockwell 3, Dineo Khabele 2, Andrew M Stern 1, Paul A Clemons 4, Alykhan F Shamji 5, Stuart L Schreiber 6
Affiliations
- PMID: 23993102
- PMCID: PMC3954635
- DOI: 10.1016/j.cell.2013.08.003
An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules
Amrita Basu et al. Cell. 2013.
Abstract
The high rate of clinical response to protein-kinase-targeting drugs matched to cancer patients with specific genomic alterations has prompted efforts to use cancer cell line (CCL) profiling to identify additional biomarkers of small-molecule sensitivities. We have quantitatively measured the sensitivity of 242 genomically characterized CCLs to an Informer Set of 354 small molecules that target many nodes in cell circuitry, uncovering protein dependencies that: (1) associate with specific cancer-genomic alterations and (2) can be targeted by small molecules. We have created the Cancer Therapeutics Response Portal (http://www.broadinstitute.org/ctrp) to enable users to correlate genetic features to sensitivity in individual lineages and control for confounding factors of CCL profiling. We report a candidate dependency, associating activating mutations in the oncogene β-catenin with sensitivity to the Bcl-2 family antagonist, navitoclax. The resource can be used to develop novel therapeutic hypotheses and to accelerate discovery of drugs matched to patients by their cancer genotype and lineage.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
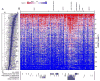
Figure 1. Response of CCLs to Informer Set
Sensitivity of 242 CCLs to small-molecule probes/drugs was assessed at dose (CellTiterGlo) and areas under the concentration-response curve (AUC) were computed. Data are shown as box plots indicating distributions of AUC values for each compound (A) and a heatmap of AUC values (scale represents AUC values ranging between 1 (sensitive; red) and 6 (unresponsive; blue)) (B) for single CCLs (columns) treated with single compounds (rows). Missing numerical values in heatmap were imputed using a k-nearest neighbors approach. AUC distributions were analyzed by incorporating context-dependent exclusions (C) of cell lines (grey bars represent excluded cell lines). See also Figure S1 and Table S1.
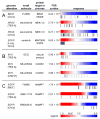
Figure 2. Genetic dependencies targeted by small molecules
The distribution of CCL response (AUC values) to compound treatment is represented as a heatmap denoting sensitivity (red) or unresponsiveness (blue) aligned with genomic alterations for corresponding CCLs (gray bars). The resource identified known clinically drug-targeted genetic dependencies (A) and known drug-resistance mechanisms (BRAF V600E outlier cell lines: *RKO; #SKMEL28). The resource also suggests dependencies with both mutation and copy number variation in MYC (B). Global analysis of the resource showed _EGFR_-mutated CCLs are unresponsive to NAMPT inhibitors (C). CNV-H: high-copy number (≥8 copies), TES: all targeted-exome sequencing mutant calls, TES-A: targeted-exome sequencing, non-neutral missense mutations; Onco: Oncomap mutant calls, MUT: any mutation call. See also Figure S2, Table S2, User Guide S1, and Table S3.
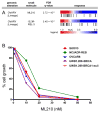
Figure 3. Lineage dependencies targeted by small molecules
Ovarian CCLs are highly sensitive to ML210 and RSL3 (A). An expanded panel of ovarian CCLs showed sensivity to ML210 (IC50 of ~10 nM) independent of the BRCA1 status of the CCLs (B). See also Figure S3.
Figure 4. Mutations in β-catenin associate with sensitivity to navitoclax
Activating mutations in β-catenin (CTNNB1) or mutations in members of its destruction complex (AXIN1; CSNK1A1) correlate with sensitivity to navitoclax (A). Previous studies have linked the Wnt/β-catenin pathway to expression of Bcl2 family members (B). An elastic-net regression model (black circles: observed; red crosses: predicted; weighted root-mean-squared error: 1.45) predicts AUC sensitivity values across CCLs treated with navitoclax (C). Heatmap depicts model features (rows; e.g., mutation, copy number) sorted by descending weight (black bars) across all CCLs tested (columns). Scale represents range of normalized values between -3 and 3 (red: relative higher copy number, presence of mutation; blue: relative lower copy number). All model features (Table S4) were input to Ingenuity Pathway Analysis (Jimenez-Marin et al., 2009) and the highest-scoring network (D) contains β-catenin as a central node (p=10−38). The network contains members of the β-catenin pathway present in the regression model (brown), other genes present in the model (dark grey), and molecular interactions with non-regression-model features (light grey). See also Table S4 and Table S5.
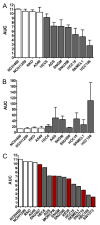
Figure 5. Confirmation experiments for navitoclax/β-catenin
Response to navitoclax observed in large-scale profiling was confirmed in 3 independent experiments (A) with the 7 most sensitive _CTNNB1-_mutant CCLs (gray bars) and 4 control CCLs lacking mutations in CTNNB1 (white bars). In parallel, caspase 3/7 activation after navitoclax treatment was measured (B), showing that loss of viability was due to induction of apotosis. The response of previously untested _CTNNB1_-mutant CCLs (red bars) to navitoclax was measured using the same conditions from our initial profiling experiments (C). Data are represented as mean +/- SD. See also Figure S4.
Figure 6. Small-molecule induction of β-catenin levels and sensitivity to navitoclax
Treatment with the GSK3β inhibitor CHIR-99021 led to increased levels of β-catenin in RKO and HT29 (non-mutant) cells (A) and relatively little change in HEC59 (non-mutant) and SW48 (S33Y CTNNB1 mutant) cells (D). Sensitivity to navitoclax was assessed after pre-treatment with CHIR-99021 (red) and compared to DMSO-pretreated controls (black). RKO (B) and HT29 (C) cells, which had increased levels of β-catenin, showed 4-fold increase in sensitivity to navitoclax, while HEC59 (E) and SW48 (F) cells, which had unchanged levels of β-catenin, demonstrate no significant change in sensitivity. Data are represented as mean +/− SD. See also Figure S5.
Similar articles
- Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset.
Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, Alexander B, Li A, Montgomery P, Wawer MJ, Kuru N, Kotz JD, Hon CS, Munoz B, Liefeld T, Dančík V, Bittker JA, Palmer M, Bradner JE, Shamji AF, Clemons PA, Schreiber SL. Seashore-Ludlow B, et al. Cancer Discov. 2015 Nov;5(11):1210-23. doi: 10.1158/2159-8290.CD-15-0235. Epub 2015 Oct 19. Cancer Discov. 2015. PMID: 26482930 Free PMC article. - More than fishing for a cure: The promises and pitfalls of high throughput cancer cell line screens.
Ling A, Gruener RF, Fessler J, Huang RS. Ling A, et al. Pharmacol Ther. 2018 Nov;191:178-189. doi: 10.1016/j.pharmthera.2018.06.014. Epub 2018 Jun 25. Pharmacol Ther. 2018. PMID: 29953899 Free PMC article. Review. - Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells.
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ. Yang W, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D955-61. doi: 10.1093/nar/gks1111. Epub 2012 Nov 23. Nucleic Acids Res. 2013. PMID: 23180760 Free PMC article. - A Road Map to Personalizing Targeted Cancer Therapies Using Synthetic Lethality.
Parameswaran S, Kundapur D, Vizeacoumar FS, Freywald A, Uppalapati M, Vizeacoumar FJ. Parameswaran S, et al. Trends Cancer. 2019 Jan;5(1):11-29. doi: 10.1016/j.trecan.2018.11.001. Epub 2018 Dec 7. Trends Cancer. 2019. PMID: 30616753 Review. - MDP, a database linking drug response data to genomic information, identifies dasatinib and statins as a combinatorial strategy to inhibit YAP/TAZ in cancer cells.
Taccioli C, Sorrentino G, Zannini A, Caroli J, Beneventano D, Anderlucci L, Lolli M, Bicciato S, Del Sal G. Taccioli C, et al. Oncotarget. 2015 Nov 17;6(36):38854-65. doi: 10.18632/oncotarget.5749. Oncotarget. 2015. PMID: 26513174 Free PMC article.
Cited by
- Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling.
Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG, Humeidi R, Peck D, Wu X, Tang AA, Wang VM, Bender SA, Lemire E, Narayan R, Montgomery P, Ben-David U, Garvie CW, Chen Y, Rees MG, Lyons NJ, McFarland JM, Wong BT, Wang L, Dumont N, O'Hearn PJ, Stefan E, Doench JG, Harrington CN, Greulich H, Meyerson M, Vazquez F, Subramanian A, Roth JA, Bittker JA, Boehm JS, Mader CC, Tsherniak A, Golub TR. Corsello SM, et al. Nat Cancer. 2020 Feb;1(2):235-248. doi: 10.1038/s43018-019-0018-6. Epub 2020 Jan 20. Nat Cancer. 2020. PMID: 32613204 Free PMC article. - Intrinsic immune evasion patterns predict temozolomide sensitivity and immunotherapy response in lower-grade gliomas.
Tu Z, Ji Q, Han Q, Long X, Li J, Wu L, Huang K, Zhu X. Tu Z, et al. BMC Cancer. 2022 Sep 12;22(1):973. doi: 10.1186/s12885-022-09984-5. BMC Cancer. 2022. PMID: 36096781 Free PMC article. - Multi-omics and pharmacological characterization of patient-derived glioma cell lines.
Wu M, Wang T, Ji N, Lu T, Yuan R, Wu L, Zhang J, Li M, Cao P, Zhao J, Li G, Li J, Li Y, Tang Y, Gao Z, Wang X, Cheng W, Ge M, Cui G, Li R, Wu A, You Y, Zhang W, Wang Q, Chen J. Wu M, et al. Nat Commun. 2024 Aug 8;15(1):6740. doi: 10.1038/s41467-024-51214-y. Nat Commun. 2024. PMID: 39112531 Free PMC article. - Conditional lethality profiling reveals anticancer mechanisms of action and drug-nutrient interactions.
Flickinger KM, Wilson KM, Rossiter NJ, Hunger AL, Vishwasrao PV, Lee TD, Mellado Fritz CA, Richards RM, Hall MD, Cantor JR. Flickinger KM, et al. Sci Adv. 2024 Oct 4;10(40):eadq3591. doi: 10.1126/sciadv.adq3591. Epub 2024 Oct 4. Sci Adv. 2024. PMID: 39365851 Free PMC article. - Gene Fusions Create Partner and Collateral Dependencies Essential to Cancer Cell Survival.
Gillani R, Seong BKA, Crowdis J, Conway JR, Dharia NV, Alimohamed S, Haas BJ, Han K, Park J, Dietlein F, He MX, Imamovic A, Ma C, Bassik MC, Boehm JS, Vazquez F, Gusev A, Liu D, Janeway KA, McFarland JM, Stegmaier K, Van Allen EM. Gillani R, et al. Cancer Res. 2021 Aug 1;81(15):3971-3984. doi: 10.1158/0008-5472.CAN-21-0791. Epub 2021 Jun 7. Cancer Res. 2021. PMID: 34099491 Free PMC article.
References
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. - PubMed
- Arteaga CL. EGF receptor mutations in lung cancer: from humans to mice and maybe back to humans. Cancer Cell. 2006;9:421–423. - PubMed
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300.
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. The Journal of biological chemistry. 2002;277:30998–31004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 CA148399/CA/NCI NIH HHS/United States
- R01 CA161061/CA/NCI NIH HHS/United States
- K08 CA148887/CA/NCI NIH HHS/United States
- RC2-CA148399/CA/NCI NIH HHS/United States
- R01 CA097061/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources