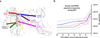Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans - PubMed (original) (raw)
. 2013 Oct;45(10):1176-82.
doi: 10.1038/ng.2744. Epub 2013 Sep 1.
Mireia Coscolla, Tao Luo, Sonia Borrell, Kathryn E Holt, Midori Kato-Maeda, Julian Parkhill, Bijaya Malla, Stefan Berg, Guy Thwaites, Dorothy Yeboah-Manu, Graham Bothamley, Jian Mei, Lanhai Wei, Stephen Bentley, Simon R Harris, Stefan Niemann, Roland Diel, Abraham Aseffa, Qian Gao, Douglas Young, Sebastien Gagneux
Affiliations
- PMID: 23995134
- PMCID: PMC3800747
- DOI: 10.1038/ng.2744
Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans
Iñaki Comas et al. Nat Genet. 2013 Oct.
Abstract
Tuberculosis caused 20% of all human deaths in the Western world between the seventeenth and nineteenth centuries and remains a cause of high mortality in developing countries. In analogy to other crowd diseases, the origin of human tuberculosis has been associated with the Neolithic Demographic Transition, but recent studies point to a much earlier origin. We analyzed the whole genomes of 259 M. tuberculosis complex (MTBC) strains and used this data set to characterize global diversity and to reconstruct the evolutionary history of this pathogen. Coalescent analyses indicate that MTBC emerged about 70,000 years ago, accompanied migrations of anatomically modern humans out of Africa and expanded as a consequence of increases in human population density during the Neolithic period. This long coevolutionary history is consistent with MTBC displaying characteristics indicative of adaptation to both low and high host densities.
Figures
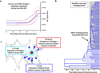
Figure 1. Genome-based phylogeny of MTBC mirrors that of human mitochondrial genomes
A, Whole-genome phylogeny of 220 strains of Mycobacterium tuberculosis complex (MTBC). Support values for the main branches after inference with Neighbour-joining (left) and Maximum-likelihood (right) are shown. B, Principal Component Analysis (PCA) of the 34,167 SNPs. The first three PCA axes are shown; these discriminate between evolutionarily “modern” (highlighted in grey) and “ancient” (all other) strains. Individual lineages are shown following the colour coding of Fig. 1A. C and D, Comparison of MTBC phylogeny (C) and a phylogeny derived from 4,955 mitochondrial genomes representative of the main human haplogroups (D). The colour coding highlights the similarities in tree topology and geographic distribution of MTBC strains and main human mitochondrial macro-haplogroups (black - African clades: MTBC Lineage 5 and 6, human mitochondrial macro-haplogroups L0-L3; pink – South-East Asian and Oceanian clades: MTBC Lineage 1, human mitochondrial macro-haplogroup M; blue – Eurasian clades: MTBC Lineage 2, 3, and 4, human mitochondrial macro-haplogroup N). The MTBC Lineage 7 has only been found in Ethiopia and its correlation with any of the three main human haplogroups remains unclear.
Figure 2. Out-of-Africa and Neolithic expansion of MTBC
A, Map summarizing the results of the phylogeographic and dating analyses of MTBC. The colour codes used for lineages are according to Fig. 1A. Major splits are annotated with the median value (in kya) of the dating of the relevant node. Lineage 7 (yellow) has so far been isolated exclusively from patients with known country of origin in the Horn of Africa. Lineage 7 diverged subsequent to the proposed Out-of-Africa migration of MTBC; it may have arisen amongst a human population that remained in Africa, or a population that returned to Africa. B, Bayesian skyline plots illustrating changes in population diversity of MTBC (red line) and humans based on mitochondrial DNA (blue lines) during the last 60 ky. Dashed lines represent the 95% highest probability density (HPD) intervals for the estimated population sizes.
Figure 3. Neolithic expansion and spread of MTBC Lineage 2 “Beijing” in East Asia
A, Bayesian skyline indicating changes in Lineage 2 diversity over time (red line) as compared to human mtDNA haplogroups from East Asia (blue line). 95% HPD intervals for the population size estimations are shown in dashed lines. B, Dated Bayesian phylogeny of the MTBC Lineage 2 based on coalescent analysis. C, Map of the parallel origin and migration of MTBC and humans in East Asia indicating the first archaeological evidence of modern human in the region 32–42 kya, coinciding with the migration of MTBC from Central to East Asia, the start of the Neolithic in the region indicated by the first evidence of domesticated crop in China coinciding with the origin of the MTBC “Beijing family” 8 kya (6–11 kya), and the co-expansion of agriculture and MTBC “Beijing family” into neighbouring countries 3–5 kya.
Figure 4. Time-dependent decay of substitution rates in bacteria based on whole-genome datasets
Scatter plot graph representing the relationship between substitution rate and time span between the most recent ancestor and the last sampling date for each studied pathogen. Values were extracted from relevant publications that use whole-genome representative datasets and coalescent analysis of substitution rates (for a complete list of references see Supplementary Table 9).
Similar articles
- A seventeenth-century Mycobacterium tuberculosis genome supports a Neolithic emergence of the Mycobacterium tuberculosis complex.
Sabin S, Herbig A, Vågene ÅJ, Ahlström T, Bozovic G, Arcini C, Kühnert D, Bos KI. Sabin S, et al. Genome Biol. 2020 Aug 10;21(1):201. doi: 10.1186/s13059-020-02112-1. Genome Biol. 2020. PMID: 32778135 Free PMC article. - Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese.
Luo T, Comas I, Luo D, Lu B, Wu J, Wei L, Yang C, Liu Q, Gan M, Sun G, Shen X, Liu F, Gagneux S, Mei J, Lan R, Wan K, Gao Q. Luo T, et al. Proc Natl Acad Sci U S A. 2015 Jun 30;112(26):8136-41. doi: 10.1073/pnas.1424063112. Epub 2015 Jun 15. Proc Natl Acad Sci U S A. 2015. PMID: 26080405 Free PMC article. - Host-pathogen coevolution in human tuberculosis.
Gagneux S. Gagneux S. Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):850-9. doi: 10.1098/rstb.2011.0316. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312052 Free PMC article. Review. - Ecology and evolution of Mycobacterium tuberculosis.
Gagneux S. Gagneux S. Nat Rev Microbiol. 2018 Apr;16(4):202-213. doi: 10.1038/nrmicro.2018.8. Epub 2018 Feb 19. Nat Rev Microbiol. 2018. PMID: 29456241 Review. - The paleopathological evidence on the origins of human tuberculosis: a review.
Buzic I, Giuffra V. Buzic I, et al. J Prev Med Hyg. 2020 Apr 30;61(1 Suppl 1):E3-E8. doi: 10.15167/2421-4248/jpmh2020.61.1s1.1379. eCollection 2020 Mar. J Prev Med Hyg. 2020. PMID: 32529097 Free PMC article. Review.
Cited by
- Genetic Lineages of Mycobacterium tuberculosis Isolates in Isfahan, Iran.
Riyahi Zaniani F, Moghim S, Mirhendi H, Ghasemian Safaei H, Fazeli H, Salehi M, Nasr Esfahani B. Riyahi Zaniani F, et al. Curr Microbiol. 2017 Jan;74(1):14-21. doi: 10.1007/s00284-016-1145-2. Epub 2016 Oct 14. Curr Microbiol. 2017. PMID: 27743106 - A seventeenth-century Mycobacterium tuberculosis genome supports a Neolithic emergence of the Mycobacterium tuberculosis complex.
Sabin S, Herbig A, Vågene ÅJ, Ahlström T, Bozovic G, Arcini C, Kühnert D, Bos KI. Sabin S, et al. Genome Biol. 2020 Aug 10;21(1):201. doi: 10.1186/s13059-020-02112-1. Genome Biol. 2020. PMID: 32778135 Free PMC article. - Post-translational Acetylation of MbtA Modulates Mycobacterial Siderophore Biosynthesis.
Vergnolle O, Xu H, Tufariello JM, Favrot L, Malek AA, Jacobs WR Jr, Blanchard JS. Vergnolle O, et al. J Biol Chem. 2016 Oct 14;291(42):22315-22326. doi: 10.1074/jbc.M116.744532. Epub 2016 Aug 26. J Biol Chem. 2016. PMID: 27566542 Free PMC article. - A smooth tubercle bacillus from Ethiopia phylogenetically close to the Mycobacterium tuberculosis complex.
Yenew B, Ghodousi A, Diriba G, Tesfaye E, Cabibbe AM, Amare M, Moga S, Alemu A, Dagne B, Sinshaw W, Mollalign H, Meaza A, Tadesse M, Gamtesa DF, Abebaw Y, Seid G, Zerihun B, Getu M, Chiacchiaretta M, Gaudin C, Marceau M, Didelot X, Tolera G, Abdella S, Kebede A, Getahun M, Mehammed Z, Supply P, Cirillo DM. Yenew B, et al. Nat Commun. 2023 Nov 18;14(1):7519. doi: 10.1038/s41467-023-42755-9. Nat Commun. 2023. PMID: 37980337 Free PMC article. - Geographic population structure and distinct intra-population dynamics of globally abundant freshwater bacteria.
Hoetzinger M, Hahn MW, Andersson LY, Buckley N, Ramsin C, Buck M, Nuy JK, Garcia SL, Puente-Sánchez F, Bertilsson S. Hoetzinger M, et al. ISME J. 2024 Jan 8;18(1):wrae113. doi: 10.1093/ismejo/wrae113. ISME J. 2024. PMID: 38959851 Free PMC article.
References
- Wilson LG. Commentary: Medicine, population, and tuberculosis. Int. J Epidemiol. 2005;34:521–524. - PubMed
- WHO G. The Global Plan to STOP TB 2011–2015. WHO. 2011
- Diamond J. Guns, Germs, and Steel: The Fates of Human Societies. Vol. 496. W. W. Norton & Company; 1999. Apr 1,
- Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449:843–849. - PubMed
Publication types
MeSH terms
Grants and funding
- AI090928 AND/AI/NIAID NIH HHS/United States
- U117581288/MRC_/Medical Research Council/United Kingdom
- 089276/WT_/Wellcome Trust/United Kingdom
- MC_U117588500/MRC_/Medical Research Council/United Kingdom
- U.1175.02.002.00015.01/MRC_/Medical Research Council/United Kingdom
- R01 AI090928/AI/NIAID NIH HHS/United States
- 098051/WT_/Wellcome Trust/United Kingdom
- MC_U117581288/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical