Genome-wide methylation analysis shows similar patterns in Barrett's esophagus and esophageal adenocarcinoma - PubMed (original) (raw)
Genome-wide methylation analysis shows similar patterns in Barrett's esophagus and esophageal adenocarcinoma
Enping Xu et al. Carcinogenesis. 2013 Dec.
Erratum in
- Carcinogenesis. 2014 Mar;35(3):738
Abstract
Barrett's esophagus (BE) is a precursor of esophageal adenocarcinoma (EAC). To identify novel tumor suppressors involved in esophageal carcinogenesis and potential biomarkers for the malignant progression of BE, we performed a genome-wide methylation profiling of BE and EAC tissues. Using Illumina's Infinium HumanMethylation27 BeadChip microarray, we examined the methylation status of 27 578 CpG sites in 94 normal esophageal (NE), 77 BE and 117 EAC tissue samples. The overall methylation of CpG sites within the CpG islands was higher, but outside of the CpG islands was lower in BE and EAC tissues than in NE tissues. Hierarchical clustering analysis showed an excellent separation of NE tissues from BE and EAC tissues; however, the clustering of BE and EAC tissues was less clear, suggesting that methylation occurs early during the progression of EAC. We confirmed many previously reported hypermethylated genes and identified a large number of novel hypermethylated genes in BE and EAC tissues, particularly genes encoding ADAM (A Disintegrin And Metalloproteinase) peptidase proteins, cadherins and protocadherins, and potassium voltage-gated channels. Pathway analysis showed that a number of channel and transporter activities were enriched for hypermethylated genes. We used pyrosequencing to validate selected candidate genes and found high correlations between the array and pyrosequencing data (rho > 0.8 for each validated gene). The differentially methylated genes and pathways may provide biological insights into the development and progression of BE and become potential biomarkers for the prediction and early detection of EAC.
Figures
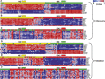
Fig. 1.
Supervised hierarchical clustering of 10 CpG sites in (A) NE tissues with EAC tissues in the discovery set, (B) NE tissues with BE tissues in the discovery set, (C) EAC with BE tissues in the discovery set, (D) NE tissues with EAC tissues in the validation set, (E) NE tissues with BE tissues in the validation set and (F) EAC with BE tissues in the validation set. Each column represents a sample and each row represents a CpG site. Methylation levels vary from fully unmethylated (blue) to fully methylated (red).
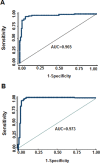
Fig. 2.
ROC curve analysis of the diagnostic efficacy of 10 common differentially methylated CpG sites in BE and EAC in all samples. (A) ROC curve for discriminating NE tissues from BE. (B) ROC curve for discriminating NE tissues from EAC. AUC, area under the ROC curve.
Similar articles
- DNA methylation profiling in Barrett's esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses.
Kaz AM, Wong CJ, Luo Y, Virgin JB, Washington MK, Willis JE, Leidner RS, Chak A, Grady WM. Kaz AM, et al. Epigenetics. 2011 Dec;6(12):1403-12. doi: 10.4161/epi.6.12.18199. Epigenetics. 2011. PMID: 22139570 Free PMC article. - Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma.
Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, Laird PW, Skinner KA. Eads CA, et al. Cancer Res. 2000 Sep 15;60(18):5021-6. Cancer Res. 2000. PMID: 11016622 - Identification of a key role of widespread epigenetic drift in Barrett's esophagus and esophageal adenocarcinoma.
Luebeck EG, Curtius K, Hazelton WD, Maden S, Yu M, Thota PN, Patil DT, Chak A, Willis JE, Grady WM. Luebeck EG, et al. Clin Epigenetics. 2017 Oct 16;9:113. doi: 10.1186/s13148-017-0409-4. eCollection 2017. Clin Epigenetics. 2017. PMID: 29046735 Free PMC article. - Cellular origins and molecular mechanisms of Barrett's esophagus and esophageal adenocarcinoma.
Fang Y, Chen X, Bajpai M, Verma A, Das KM, Souza RF, Garman KS, Donohoe CL, O'Farrell NJ, Reynolds JV, Dvorak K. Fang Y, et al. Ann N Y Acad Sci. 2013 Oct;1300:187-199. doi: 10.1111/nyas.12249. Ann N Y Acad Sci. 2013. PMID: 24117642 Review. - Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma.
Agarwal A, Polineni R, Hussein Z, Vigoda I, Bhagat TD, Bhattacharyya S, Maitra A, Verma A. Agarwal A, et al. Int J Clin Exp Pathol. 2012;5(5):382-96. Epub 2012 May 23. Int J Clin Exp Pathol. 2012. PMID: 22808291 Free PMC article. Review.
Cited by
- Kyoto international consensus report on anatomy, pathophysiology and clinical significance of the gastro-oesophageal junction.
Sugano K, Spechler SJ, El-Omar EM, McColl KEL, Takubo K, Gotoda T, Fujishiro M, Iijima K, Inoue H, Kawai T, Kinoshita Y, Miwa H, Mukaisho KI, Murakami K, Seto Y, Tajiri H, Bhatia S, Choi MG, Fitzgerald RC, Fock KM, Goh KL, Ho KY, Mahachai V, O'Donovan M, Odze R, Peek R, Rugge M, Sharma P, Sollano JD, Vieth M, Wu J, Wu MS, Zou D, Kaminishi M, Malfertheiner P. Sugano K, et al. Gut. 2022 Aug;71(8):1488-1514. doi: 10.1136/gutjnl-2022-327281. Epub 2022 Jun 20. Gut. 2022. PMID: 35725291 Free PMC article. - Integrated molecular analysis reveals complex interactions between genomic and epigenomic alterations in esophageal adenocarcinomas.
Peng D, Guo Y, Chen H, Zhao S, Washington K, Hu T, Shyr Y, El-Rifai W. Peng D, et al. Sci Rep. 2017 Jan 19;7:40729. doi: 10.1038/srep40729. Sci Rep. 2017. PMID: 28102292 Free PMC article. - Increased Expression of Kv10.2 in the Hippocampus Attenuates Valproic Acid-Induced Autism-Like Behaviors in Rats.
Wang J, Feng S, Li M, Liu Y, Yan J, Tang Y, Du D, Chen F. Wang J, et al. Neurochem Res. 2019 Dec;44(12):2796-2808. doi: 10.1007/s11064-019-02903-4. Epub 2019 Nov 15. Neurochem Res. 2019. PMID: 31728858 - Barrett's esophagus: The pathomorphological and molecular genetic keystones of neoplastic progression.
Maslyonkina KS, Konyukova AK, Alexeeva DY, Sinelnikov MY, Mikhaleva LM. Maslyonkina KS, et al. Cancer Med. 2022 Jan;11(2):447-478. doi: 10.1002/cam4.4447. Epub 2021 Dec 6. Cancer Med. 2022. PMID: 34870375 Free PMC article. Review. - From genetics to signaling pathways: molecular pathogenesis of esophageal adenocarcinoma.
Caspa Gokulan R, Garcia-Buitrago MT, Zaika AI. Caspa Gokulan R, et al. Biochim Biophys Acta Rev Cancer. 2019 Aug;1872(1):37-48. doi: 10.1016/j.bbcan.2019.05.003. Epub 2019 May 30. Biochim Biophys Acta Rev Cancer. 2019. PMID: 31152823 Free PMC article. Review.
References
- Siegel R., et al. (2013) Cancer statistics, 2013. CA. Cancer J. Clin., 63, 11–30 - PubMed
- Montgomery E., et al. (2001). Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum. Pathol., 32, 368–378 - PubMed
- Hongo M., et al. (2009). Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J. Gastroenterol. Hepatol., 24, 729–735 - PubMed
- Wild C.P., et al. (2003). Reflux, Barrett’s oesophagus and adenocarcinoma: burning questions. Nat. Rev. Cancer, 3, 676–684 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases