The head module of Mediator directs activation of preloaded RNAPII in vivo - PubMed (original) (raw)
The head module of Mediator directs activation of preloaded RNAPII in vivo
Sarah K Lee et al. Nucleic Acids Res. 2013 Dec.
Abstract
The successful synthesis of a transcript by RNA polymerase II (RNAPII) is a multistage process with distinct rate-limiting steps that can vary depending on the particular gene. A growing number of genes in a variety of organisms are regulated at steps after the recruitment of RNAPII. The best-characterized Saccharomyces cerevisiae gene regulated in this manner is CYC1. This gene has high occupancy of RNAPII under non-inducing conditions, defining it as a poised gene. Here, we find that subunits of the head module of Mediator, Med18 and Med20, and Med19 are required for activation of transcription at the CYC1 promoter in response to environmental cues. These subunits of Mediator are required at the preloaded promoter for normal levels of recruitment and activity of the general transcription factor TFIIH. Strikingly, these Mediator components are dispensable for activation by the same activator at a different gene, which lacks a preloaded polymerase in the promoter region. Based on these results and other studies, we speculate that Mediator plays an essential role in triggering an inactive polymerase at CYC1 into a productively elongating form.
Figures
Figure 1.
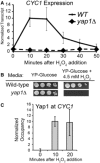
Yap1 directly regulates CYC1 expression during oxidative stress. (A) CYC1 transcript levels are rapidly induced in response to oxidative stress. An S1 nuclease protection assay was performed with RNA isolated from wild-type and yap1Δ cells before exposure to H2O2 and in 10-min intervals after adding H2O2 (0.3 mM). A probe for tRNAw was used as a loading control, and the transcript level in the wild-type strain 10 min after H2O2 exposure was set to 10. Results represent the average of at least three biological replicates ±SD. (B) Serial spot dilutions of the wild-type and yap1Δ strains on YP-Glucose and on YP-Glucose containing H2O2 (4.5 mM). Plates were incubated at 30°C for 3 days before photographing. (C) Yap1-myc occupies the CYC1 promoter during oxidative stress. Occupancy was determined with a ChIP using a Yap1-myc strain. Formaldehyde was added to cross-link proteins and DNA at the time point indicated on the _x_-axis. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
Figure 2.
Activator dependence at CYC1. (A) CYC1 activation during growth in non-fermentable carbon sources is Hap4-dependent. An S1 nuclease protection assay was used to analyze RNA prepared from the wild-type, _yap1_Δ and _hap4_Δ strains after cells were transferred to media containing 3% ethanol as the carbon source. A tRNAw probe was used as a loading control. Representative gel is shown with quantification of transcript levels 6 h after transfer to a non-fermentable carbon source. Levels were analyzed in biological triplicate. (B) CYC1 activation during oxidative stress is Yap1-dependent. Transcript levels were analyzed as described earlier in the text with RNA collected 10, 20 and 30 min after the addition of H2O2 to growth media. A tRNAw probe was used as a loading control in the S1 assays. Representative gel is shown with quantification of CYC1 transcript levels in each strain 10 min after the addition of H2O2. Levels were analyzed in biological triplicate.
Figure 3.
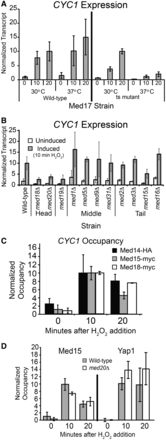
CYC1 expression is Mediator dependent during oxidative stress. (A) CYC1 expression in strains with wild-type Med17 and a temperature sensitive mutant of Med17 (srb4-138). Transcript levels are shown at the permissive (30°C) and non-permissive temperature (37°C) in the uninduced condition, and 10 and 20 min after the addition of H2O2, as indicated. CYC1 transcript levels were normalized to a tRNAw probe, expression level in the wild-type strain 20 min after activation was set to 10. The average ±SD of three biological replicates is shown. (B) Certain non-essential subunits of the Mediator complex are required for CYC1 activation during oxidative stress. Transcript levels were analyzed with RNA prepared from the wild-type strain and strains containing deletions of the indicated Mediator subunit in both uninduced (YP-Glucose, unfilled bars) and induced (10 min H2O2 treatment, gray bars) conditions. CYC1 transcript level was normalized as described earlier in the text. Expression in the wild-type strain at 10 min after activation was set to 10. The average ±SD of three biological replicates is shown for all strains except the tail module deletions, which is shown in biological duplicate. (C) Mediator subunits are recruited to CYC1 during oxidative stress. Occupancy of Med14-HA, Med15-myc and Med18-myc was determined with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of two or three biological replicates. (D) Mediator and Yap1 occupancy at the CYC1 promoter is independent of Med20, as occupancy of Med15-myc and Yap1-myc increases on gene activation in both the wild-type (gray bars) and med20Δ strain (white bars). Occupancy of Med15-myc and Yap1-myc was determined with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
Figure 4.
MED18, MED19 and MED20 are required during oxidative stress and growth on non-fermentable carbon sources. Serial spot dilutions of the wild-type strain and strains containing deletions of 11 Mediator subunits on YP-Glucose, YP-Glucose supplemented with H2O2 and YP-Ethanol/Glycerol. Plates were incubated at 30_°_C for 2–5 days before photographing.
Figure 5.
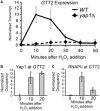
GTT2 is a Yap1-dependent gene without preloaded RNAPII. (A) GTT2 transcript levels are rapidly induced in response to oxidative stress. An S1 nuclease protection assay was performed with RNA isolated from wild-type and yap1Δ cells before exposure to H2O2 and in 10-min intervals after adding H2O2 (0.3 mM). A probe for tRNAw was used as a loading control, and the transcript level in the wild-type strain 10 min after H2O2 exposure was set to 10. Points represent the average ±SD of at least three biological replicates for the wild-type, and in biological duplicate for the yap1Δ strain. (B) Yap1 directly stimulates GTT2 transcription, as Yap1-myc occupies this promoter during oxidative stress. Occupancy was determined with a ChIP of a Yap1-myc fusion protein. Occupancy at the GAL10 promoter region was subtracted from the occupancy at GTT2, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates. (C) RNAPII does not occupy the GTT2 promoter before gene activation but is recruited on activation. Occupancy at the GAL10 promoter region was subtracted from the occupancy at GTT2, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
Figure 6.
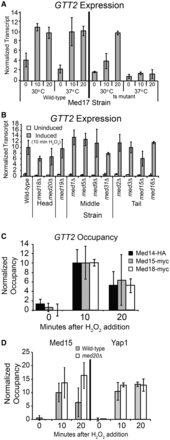
GTT2 activation is Mediator dependent, but does not rely on the Med18, Med20 and Med20 subunits for expression. (A) GTT2 expression in strains with wild-type Med17 and a temperature sensitive mutant of Med17 (srb4-138). Transcript levels are shown at the permissive (30°C) and non-permissive temperature (37°C) in the uninduced condition, and 10 and 20 min after the addition of H2O2, as indicated. Transcript levels were analyzed as described in Figure 3. The average ±SD of three biological replicates is shown. (B) GTT2 activation is largely independent of the Med18, Med19 and Med20 proteins. RNA levels from indicated strains were analyzed as described in Figure 3. The average ±SD of three biological replicates is shown for all strains except the tail module deletions, which is shown in biological duplicate. (C) Mediator subunits are recruited to GTT2 during oxidative stress. Occupancy of Med14-HA, Med15-myc and Med18-myc were determined with a time course ChIP and analyzed as described in Figure 3. (D). Mediator and Yap1 occupancy at the GTT2 promoter is independent of Med20, as occupancy of Med15-myc and Yap1-myc increases on gene activation in both the wild-type (gray bars) and med20Δ strain (white bars). Occupancy of Med15-myc and Yap1-myc was determined with a time course ChIP and analyzed as described in Figure 3.
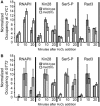
Figure 7.
Med20 is required for normal TFIIH recruitment and Ser5 phosphorylation at the promoter of CYC1 but not at GTT2. (A) Occupancy of RNAPII, Kin28-myc, Ser5-P and Rad3-HA in the wild-type (gray bars) and _med20_Δ (white bars) strains was determined at the CYC1 promoter with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point in the wild-type strain was set to 10. Bars represent the average ±SD of three biological replicates. (B) Occupancy of RNAPII, Kin28-myc, Ser5-P and Rad3-HA in the wild-type (gray bars) and _med20_Δ (white bars) strains was determined at the GTT2 promoter as described in (A).
Similar articles
- Activation of a poised RNAPII-dependent promoter requires both SAGA and mediator.
Lee SK, Fletcher AG, Zhang L, Chen X, Fischbeck JA, Stargell LA. Lee SK, et al. Genetics. 2010 Mar;184(3):659-72. doi: 10.1534/genetics.109.113464. Epub 2010 Jan 4. Genetics. 2010. PMID: 20048049 Free PMC article. - Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II.
Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. Zhang L, et al. Mol Cell Biol. 2008 Feb;28(4):1393-403. doi: 10.1128/MCB.01733-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086892 Free PMC article. - A Role for Mediator Core in Limiting Coactivator Recruitment in Saccharomyces cerevisiae.
Yarrington RM, Yu Y, Yan C, Bai L, Stillman DJ. Yarrington RM, et al. Genetics. 2020 Jun;215(2):407-420. doi: 10.1534/genetics.120.303254. Epub 2020 Apr 23. Genetics. 2020. PMID: 32327563 Free PMC article. - Origins and activity of the Mediator complex.
Conaway RC, Conaway JW. Conaway RC, et al. Semin Cell Dev Biol. 2011 Sep;22(7):729-34. doi: 10.1016/j.semcdb.2011.07.021. Epub 2011 Jul 28. Semin Cell Dev Biol. 2011. PMID: 21821140 Free PMC article. Review. - The yeast Mediator complex and its regulation.
Björklund S, Gustafsson CM. Björklund S, et al. Trends Biochem Sci. 2005 May;30(5):240-4. doi: 10.1016/j.tibs.2005.03.008. Trends Biochem Sci. 2005. PMID: 15896741 Review.
Cited by
- Med15B Regulates Acid Stress Response and Tolerance in Candida glabrata by Altering Membrane Lipid Composition.
Qi Y, Liu H, Yu J, Chen X, Liu L. Qi Y, et al. Appl Environ Microbiol. 2017 Aug 31;83(18):e01128-17. doi: 10.1128/AEM.01128-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28710262 Free PMC article. - CgMED3 Changes Membrane Sterol Composition To Help Candida glabrata Tolerate Low-pH Stress.
Lin X, Qi Y, Yan D, Liu H, Chen X, Liu L. Lin X, et al. Appl Environ Microbiol. 2017 Aug 17;83(17):e00972-17. doi: 10.1128/AEM.00972-17. Print 2017 Sep 1. Appl Environ Microbiol. 2017. PMID: 28667115 Free PMC article. - Candida glabrata Med3 Plays a Role in Altering Cell Size and Budding Index To Coordinate Cell Growth.
Liu H, Kong L, Qi Y, Chen X, Liu L. Liu H, et al. Appl Environ Microbiol. 2018 Jul 17;84(15):e00781-18. doi: 10.1128/AEM.00781-18. Print 2018 Aug 1. Appl Environ Microbiol. 2018. PMID: 29776932 Free PMC article. - Transcriptomic Insights into the Effect of Melatonin in Saccharomyces cerevisiae in the Presence and Absence of Oxidative Stress.
Sunyer-Figueres M, Vázquez J, Mas A, Torija MJ, Beltran G. Sunyer-Figueres M, et al. Antioxidants (Basel). 2020 Oct 1;9(10):947. doi: 10.3390/antiox9100947. Antioxidants (Basel). 2020. PMID: 33019712 Free PMC article. - Oxidant-Sensing Pathways in the Responses of Fungal Pathogens to Chemical Stress Signals.
Simaan H, Lev S, Horwitz BA. Simaan H, et al. Front Microbiol. 2019 Mar 19;10:567. doi: 10.3389/fmicb.2019.00567. eCollection 2019. Front Microbiol. 2019. PMID: 30941117 Free PMC article. Review.
References
- Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 2010;79:271–293. - PubMed
- Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous