Type IV pili in Gram-positive bacteria - PubMed (original) (raw)
Review
Type IV pili in Gram-positive bacteria
Stephen Melville et al. Microbiol Mol Biol Rev. 2013 Sep.
Abstract
Type IV pili (T4P) are surface-exposed fibers that mediate many functions in bacteria, including locomotion, adherence to host cells, DNA uptake (competence), and protein secretion and that can act as nanowires carrying electric current. T4P are composed of a polymerized protein, pilin, and their assembly apparatuses share protein homologs with type II secretion systems in eubacteria and the flagella of archaea. T4P are found throughout Gram-negative bacterial families and have been studied most extensively in certain model Gram-negative species. Recently, it was discovered that T4P systems are also widespread among Gram-positive species, in particular the clostridia. Since Gram-positive and Gram-negative bacteria have many differences in cell wall architecture and other features, it is remarkable how similar the T4P core proteins are between these organisms, yet there are many key and interesting differences to be found as well. In this review, we compare the two T4P systems and identify and discuss the features they have in common and where they differ to provide a very broad-based view of T4P systems across all eubacterial species.
Figures
Fig 1
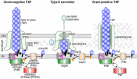
Illustration of key components in the Gram-negative T4P and T2S systems and the Gram-positive T4P system and their localization in the bacterial envelope. The descriptive name for each component is shown for the Gram-negative T4P system, and common names for each component are listed for all three systems. The inner membrane core protein (IMCP), the assembly ATPase, inner membrane accessory proteins (not shown), and possibly the retraction ATPase form the inner membrane pilus assembly platform. The cylinder surrounding the pilus in the Gram-positive T4P illustration represents a hypothetical protein channel, through which the pilus extends and retracts, transiting the peptidoglycan layer. IM, inner membrane; OM, outer membrane.
Fig 2
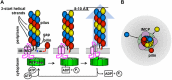
Model for T4P assembly. (A) Side view of the inner membrane assembly platform. In the pilus assembly cycle shown, a single pilin subunit docks into a gap in the growing pilus filament, attracted by complementarity between its negatively charged Glu5 (−) residue and the positively charged main chain amine on the N-terminal residue (+) of the terminal pilin subunit in the growing filament. ATP is hydrolyzed by the assembly ATPase, inducing a conformational change in the inner membrane core protein (IMCP) that extrudes the filament outward a short distance, opening up a new gap ∼120° around the base of the filament for a new pilin subunit to dock. Thus, subunits are added iteratively at 3 sites around the base of the growing filament, corresponding to each of the three helical strands in the T4P, shown in red, blue, and yellow. Each subunit is staggered by an axial distance of 8 to 10 Å along the length of the filament. Only one of three predicted inner membrane core proteins is shown. IM, inner membrane. (B) Top view of the assembly apparatus, looking down on the growing filament. The red subunit is added first, followed by the blue, then the yellow, etc. (Modified from reference with permission from Elsevier.)
Fig 3
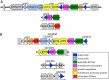
T4P gene clusters in C. perfringens and C. difficile. The primary T4P operon is shown at the top for C. perfringens strain 13 (A) and C. difficile strain 630 (B), and the secondary gene clusters are shown below. Protein annotations are based on sequence homology with proteins of known function, gene organization, and functional analysis. Genes are colored the same as their protein products shown in Fig. 1 and 2.
Fig 4
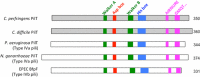
Amino acid sequence similarity among C. perfringens pilins. (A) Amino acid sequences of the signal peptides and N-terminal 30 residues of the mature pilins from C. perfringens strain 13. The red background indicates amino acid sequence identity; the yellow background and bold letters indicate sequence similarity (nonbolded amino acids within each yellow column are not conserved). Residues are numbered above the sequences: negative numbers are used for the signal peptide, and positive numbers are used for the mature pilin sequence. (B) Schematic comparison of C. perfringens strain 13 pilins and putative pilins. Asterisks indicate putative minor pilins, among which CPE2279 is a GspK family member based on its large size and a tyrosine at position 5. The identities of the first and fifth amino acids of the mature protein are indicated. Gray bars represent the signal peptide, and yellow bars indicate amino acid sequence homology among the pilins. (C) Schematic comparison of pilins among multiple C. perfringens strains. Red bars indicate sequence identity for the majority of the residues among different strains, and blue bars indicate amino acid insertions/differences.
Fig 5
Amino acid sequence similarity among C. difficile putative type IV pilins. (A) Amino acid sequences of the signal peptides and N-terminal 30 residues of the mature pilins from C. difficile strain 630. The pilins are listed as they appear in the operons in Fig. 3B. The red background indicates amino acid sequence identity; the yellow background and bold letters indicate sequence similarity (nonbolded amino acids within yellow columns are not conserved). Residues are numbered above the sequences: negative numbers are used for the signal peptide, and positive numbers are used for the mature pilin sequence. (B) Schematic comparison of C. difficile strain 13 pilins and putative pilins. Asterisks indicate putative minor pilins, among which 3506 is a GspK family member based on its large size and a leucine at position 5. The identities of the first and fifth amino acids of the mature protein are indicated, as are characteristic motifs described in the text. Gray bars represent the signal peptide, and yellow bars indicate amino acid sequence homology among the pilins within this strain.
Fig 6
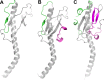
Structural predictions for Clostridium pilins. (A) Predictive model for C. perfringens strain 13 PilA1 residues 1 to 115 (of the 117-residue protein), based on the D. nodosus FimA structure (PDB ID 3SOK) (33) (17% sequence identity). The model was derived from the Phyre2 server, with 99.9% confidence. (B) Predictive model for C. perfringens strain 13 PilA2 residues 1 to 142 (of the 170-residue protein), based on the D. nodosus FimA structure (21% sequence identity). The model was derived from the Fugue server, with 99% confidence. (C) Predictive model for C. difficile strain 630 CD3513 residues 1 to 162 (of the 162-residue protein), based on the N. gonorrhoeae GC pilin (17% sequence identity). The model was derived from the Fugue server, with 95% confidence. The N-terminal α-helix and core β-sheet are shown in gray. The αβ-loop and D-region are colored green and magenta, respectively, based on their positions in the template structure.
Fig 7
Schematic of clostridial PilD prepilin peptidases. The bars show a schematic alignment of PilD homologs from clostridia and representative T4P and T2S systems. Proteins are listed based on the alignment shown in Fig. S3 in the supplemental material, with the clostridial peptidases shown in gray. The two active site aspartates are indicated, the second of which lies within the conserved GXGD motif. The schematics shown are for N. gonorrhoeae strain MS11 PilD (GenBank accession number ZP_06134023), Klebsiella oxytoca KCTC1686 PilD (YP_005018216), P. aeruginosa PAO1 PilD (NP_253218), C. difficile 630 PilD1 (YP_001090024) and PilD2 (YP_001090025), C. perfringens 13 PilD (NP_563203), V. cholerae N16961 TcpJ (NP_230487), and EPEC B171 BfpP (NP_053073).
Fig 8
Schematics of PilB assembly ATPases. Proteins are listed based on the alignment shown in Fig. S4 in the supplemental material, with the clostridial sequences shown in gray. Colored boxes delineate motifs characteristic of secretion family ATPases. Schematics are shown for V. cholerae strain N16961 EpsE (GenBank accession number NP_232359), K. oxytoca KCTC1686 PulE (YP_005018226), P. aeruginosa PAO1 PilB (NP_253216), N. gonorrhoeae MS11 PilF (P37094), C. perfringens 13 PilB2 (NP_563202) and PilB1 (NP_562760), C. difficile 630 PilB2 (YP_001090033) and PilB1 (ZP_17077753), V. cholerae N16961 TcpT (NP_230483), and EPEC B171 BfpD (NP_053070).
Fig 9
Alignment of PilC inner membrane core proteins. Proteins are listed based on the alignment shown in Fig. S5 in the supplemental material, with the clostridial sequences shown in gray. Absolutely conserved residues are indicated. These correspond to Glu82, Gly85, Pro115, and Pro135 in V. cholerae TcpE. The predicted membrane topology is shown for V. cholerae TcpE, as follows: cyto1 and cyto2, cytoplasmic domains (yellow); TM1, TM2, and TM3, transmembrane domains (gray); and peri, periplasmic loop of ∼30 amino acids (orange). Schematics are shown for P. aeruginosa strain PAO1 PilC (GenBank accession number P22609), N. gonorrhoeae MS11 PilG (AAC43469), C. perfringens 13 PilC2 (NP_563201.1) and PilC1 (NP_562759), C. difficile 630 PilC2 (YP_001090032) and PilC1 (YP_001089811), V. cholerae N16961 TcpE (P0C6C9) and EpsF (NP_232358), K. oxytoca KCTC1686 PulF (YP_005018164), and EPEC B171 BfpE (BAA84844).
Fig 10
Membrane topologies of PilM, PilN, and PilO proteins and homologs in the clostridial, type IVa, type IVb, and T2S systems. PilM is shown in red, PilN in blue, and PilO in green. Transmembrane accessory proteins with both a cytoplasmic domain and a periplasmic domain are shown in purple to suggest a hybrid PilM/PilN protein. The number of amino acid residues for each protein is indicated in parentheses. For the proteins in the Gram-positive Clostridium species, the “periplasm” refers to the region between the cytoplasmic membrane and the peptidoglycan layer. Topology predictions are shown for C. perfringens 13 PilM (GenBank accession number NP_563199), C. perfringens 13 PilN (NP_563198), C. perfringens 13 PilO (NP_563197), C. difficile 603 PilM2 (YP_001090031), C. difficile 630 PilM1 (YP_001089809), and C. difficile 630 PilO (YP_001090030).
Fig 11
Alignment of PilT retraction ATPases. Colored boxes delineate motifs characteristic of as well as unique to retraction family ATPases, with the clostridial sequences shown in gray. Schematics are shown for C. perfringens PilT (GenBank accession number NP_562683), C. difficile PilT (YP_001090026), P. aeruginosa PilT (NP_249086), N. gonorrhoeae PilT (ZP_06134424), and EPEC BfpF (ZP_03062138).
Similar articles
- Motility and adhesion through type IV pili in Gram-positive bacteria.
Piepenbrink KH, Sundberg EJ. Piepenbrink KH, et al. Biochem Soc Trans. 2016 Dec 15;44(6):1659-1666. doi: 10.1042/BST20160221. Biochem Soc Trans. 2016. PMID: 27913675 Free PMC article. Review. - Type IV pili: dynamics, biophysics and functional consequences.
Craig L, Forest KT, Maier B. Craig L, et al. Nat Rev Microbiol. 2019 Jul;17(7):429-440. doi: 10.1038/s41579-019-0195-4. Nat Rev Microbiol. 2019. PMID: 30988511 Review. - The Biosynthesis and Structures of Bacterial Pili.
Lukaszczyk M, Pradhan B, Remaut H. Lukaszczyk M, et al. Subcell Biochem. 2019;92:369-413. doi: 10.1007/978-3-030-18768-2_12. Subcell Biochem. 2019. PMID: 31214993 Review. - A tale of two pili: assembly and function of pili in bacteria.
Kline KA, Dodson KW, Caparon MG, Hultgren SJ. Kline KA, et al. Trends Microbiol. 2010 May;18(5):224-32. doi: 10.1016/j.tim.2010.03.002. Epub 2010 Apr 8. Trends Microbiol. 2010. PMID: 20378353 Free PMC article. Review. - Type IV pilin proteins: versatile molecular modules.
Giltner CL, Nguyen Y, Burrows LL. Giltner CL, et al. Microbiol Mol Biol Rev. 2012 Dec;76(4):740-72. doi: 10.1128/MMBR.00035-12. Microbiol Mol Biol Rev. 2012. PMID: 23204365 Free PMC article. Review.
Cited by
- Role of RNase Y in Clostridium perfringens mRNA Decay and Processing.
Obana N, Nakamura K, Nomura N. Obana N, et al. J Bacteriol. 2016 Dec 28;199(2):e00703-16. doi: 10.1128/JB.00703-16. Print 2017 Jan 15. J Bacteriol. 2016. PMID: 27821608 Free PMC article. - Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile.
Maldarelli GA, De Masi L, von Rosenvinge EC, Carter M, Donnenberg MS. Maldarelli GA, et al. Pathog Dis. 2014 Aug;71(3):302-14. doi: 10.1111/2049-632X.12137. Epub 2014 Feb 18. Pathog Dis. 2014. PMID: 24550179 Free PMC article. - Electroactivity across the cell wall of Gram-positive bacteria.
Paquete CM. Paquete CM. Comput Struct Biotechnol J. 2020 Nov 21;18:3796-3802. doi: 10.1016/j.csbj.2020.11.021. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33335679 Free PMC article. Review. - Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile.
Piepenbrink KH, Maldarelli GA, Martinez de la Peña CF, Dingle TC, Mulvey GL, Lee A, von Rosenvinge E, Armstrong GD, Donnenberg MS, Sundberg EJ. Piepenbrink KH, et al. Structure. 2015 Feb 3;23(2):385-96. doi: 10.1016/j.str.2014.11.018. Epub 2015 Jan 15. Structure. 2015. PMID: 25599642 Free PMC article. - The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle.
Nelson WC, Stegen JC. Nelson WC, et al. Front Microbiol. 2015 Jul 21;6:713. doi: 10.3389/fmicb.2015.00713. eCollection 2015. Front Microbiol. 2015. PMID: 26257709 Free PMC article.
References
- Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 - PubMed
- Burrows LL. 2012. Prime time for minor subunits of the type II secretion and type IV pilus systems. Mol. Microbiol. 86:765–769 - PubMed
- Burrows LL. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888 - PubMed
- Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources