Infernal 1.1: 100-fold faster RNA homology searches - PubMed (original) (raw)
Infernal 1.1: 100-fold faster RNA homology searches
Eric P Nawrocki et al. Bioinformatics. 2013.
Abstract
Summary: Infernal builds probabilistic profiles of the sequence and secondary structure of an RNA family called covariance models (CMs) from structurally annotated multiple sequence alignments given as input. Infernal uses CMs to search for new family members in sequence databases and to create potentially large multiple sequence alignments. Version 1.1 of Infernal introduces a new filter pipeline for RNA homology search based on accelerated profile hidden Markov model (HMM) methods and HMM-banded CM alignment methods. This enables ∼100-fold acceleration over the previous version and ∼10 000-fold acceleration over exhaustive non-filtered CM searches.
Availability: Source code, documentation and the benchmark are downloadable from http://infernal.janelia.org. Infernal is freely licensed under the GNU GPLv3 and should be portable to any POSIX-compliant operating system, including Linux and Mac OS/X. Documentation includes a user's guide with a tutorial, a discussion of file formats and user options and additional details on methods implemented in the software.
Contact: nawrockie@janelia.hhmi.org
Figures
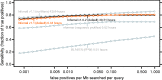
Fig. 1.
ROC-like curves for the benchmark. Plots are shown for the new Infernal 1.1 with and without filters, for the old Infernal 1.0.2, for profile HMM searches with nhmmer (from the HMMER package included in Infernal 1.1, default parameters) and for family-pairwise-searches with BLASTN (ncbi-blast-2.2.28+, default parameters). The maximum sensitivity (not shown) for default Infernal 1.1 is 0.81 (629 of 820 true positives found), which is achieved at a false-positive rate of 0.19/Mb/query. For non-filtered Infernal, maximum sensitivity is 0.87 at 2.9 false positives per Mb per query. This indicates that at high false-positive rates the filters prevent some true positives from being found, but prevent many more false positives from being found. CPU times are total times for all 106 family searches measured for single execution threads on 3.0 GHz Intel Xeon processors. The Infernal times do not include time required for model calibration.
Similar articles
- Infernal 1.0: inference of RNA alignments.
Nawrocki EP, Kolbe DL, Eddy SR. Nawrocki EP, et al. Bioinformatics. 2009 May 15;25(10):1335-7. doi: 10.1093/bioinformatics/btp157. Epub 2009 Mar 23. Bioinformatics. 2009. PMID: 19307242 Free PMC article. - nhmmer: DNA homology search with profile HMMs.
Wheeler TJ, Eddy SR. Wheeler TJ, et al. Bioinformatics. 2013 Oct 1;29(19):2487-9. doi: 10.1093/bioinformatics/btt403. Epub 2013 Jul 9. Bioinformatics. 2013. PMID: 23842809 Free PMC article. - Query-dependent banding (QDB) for faster RNA similarity searches.
Nawrocki EP, Eddy SR. Nawrocki EP, et al. PLoS Comput Biol. 2007 Mar 30;3(3):e56. doi: 10.1371/journal.pcbi.0030056. Epub 2007 Feb 7. PLoS Comput Biol. 2007. PMID: 17397253 Free PMC article. - Computational identification of functional RNA homologs in metagenomic data.
Nawrocki EP, Eddy SR. Nawrocki EP, et al. RNA Biol. 2013 Jul;10(7):1170-9. doi: 10.4161/rna.25038. Epub 2013 May 20. RNA Biol. 2013. PMID: 23722291 Free PMC article. Review. - The art of editing RNA structural alignments.
Andersen ES. Andersen ES. Methods Mol Biol. 2014;1097:379-94. doi: 10.1007/978-1-62703-709-9_17. Methods Mol Biol. 2014. PMID: 24639168 Review.
Cited by
- Intronic RNA secondary structural information captured for the human MYC pre-mRNA.
Eich TO, O'Leary CA, Moss WN. Eich TO, et al. NAR Genom Bioinform. 2024 Oct 24;6(4):lqae143. doi: 10.1093/nargab/lqae143. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39450312 Free PMC article. - Hepatitis delta virus-like circular RNAs from diverse metazoans encode conserved hammerhead ribozymes.
de la Peña M, Ceprián R, Casey JL, Cervera A. de la Peña M, et al. Virus Evol. 2021 Feb 18;7(1):veab016. doi: 10.1093/ve/veab016. eCollection 2021 Jan. Virus Evol. 2021. PMID: 33708415 Free PMC article. - The miniature genome of broad mite, Polyphagotarsonemus latus (Tarsonemidae: Acari).
Mohan M, Augustine N, Selvamani SB, P J A, Selvapandian U, Pathak J, Gracy R G, Thiruvengadam V, S N S. Mohan M, et al. Sci Data. 2024 Jul 9;11(1):748. doi: 10.1038/s41597-024-03579-4. Sci Data. 2024. PMID: 38982074 Free PMC article. - High Diversity and Functional Potential of Undescribed "Acidobacteriota" in Danish Wastewater Treatment Plants.
Kristensen JM, Singleton C, Clegg LA, Petriglieri F, Nielsen PH. Kristensen JM, et al. Front Microbiol. 2021 Apr 22;12:643950. doi: 10.3389/fmicb.2021.643950. eCollection 2021. Front Microbiol. 2021. PMID: 33967982 Free PMC article. - The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum.
King R, Urban M, Hammond-Kosack MC, Hassani-Pak K, Hammond-Kosack KE. King R, et al. BMC Genomics. 2015 Jul 22;16(1):544. doi: 10.1186/s12864-015-1756-1. BMC Genomics. 2015. PMID: 26198851 Free PMC article.
References
- Brown MP. Small subunit ribosomal RNA modeling using stochastic context-free grammars. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:57–66. - PubMed
- Durbin R, et al. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge University Press; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources