Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients - PubMed (original) (raw)
Clinical Trial
. 2013 Oct;13(10):2619-33.
doi: 10.1111/ajt.12424. Epub 2013 Sep 6.
L Chen, R K Shields, Y Zhao, S Cheng, K D Chavda, B Hao, J H Hong, Y Doi, E J Kwak, F P Silveira, R Abdel-Massih, T Bogdanovich, A Humar, D S Perlin, B N Kreiswirth, M Hong Nguyen
Affiliations
- PMID: 24011185
- PMCID: PMC3955300
- DOI: 10.1111/ajt.12424
Clinical Trial
Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients
C J Clancy et al. Am J Transplant. 2013 Oct.
Abstract
We conducted a retrospective study of 17 transplant recipients with carbapenem-resistant Klebsiella pneumoniae bacteremia, and described epidemiology, clinical characteristics and strain genotypes. Eighty-eight percent (15/17) of patients were liver or intestinal transplant recipients. Outcomes were death due to septic shock (18%), cure (24%) and persistent (>7 days) or recurrent bacteremia (29% each). Thirty- and 90-day mortality was 18% and 47%, respectively. Patients who were cured received at least one active antimicrobial agent and underwent source control interventions. Forty-one percent (7/17) of patients had intra-abdominal infections; all except one developed persistent/recurrent bacteremia despite drainage. Two patients tolerated persistent bacteremia for >300 days. All patients except one were infected with sequence type 258 (ST258), K. pneumoniae carbapenemase (KPC)-2-producing strains harboring a mutant ompK35 porin gene; the exception was infected with an ST37, KPC-3-producing strain. Seventy-one percent (12/17) of patients were infected with ST258 ompK36 mutant strains. In two patients, persistent bacteremia was caused by two strains with different ompK36 genotypes. Three ompK36 mutations were associated with significantly higher carbapenem minimum inhibitory concentrations than wild-type ompK36. Pulse-field gel electrophoresis identified a single ST258 lineage; serial strains from individual patients were indistinguishable. In conclusion, KPC-K. pneumoniae bacteremia exhibited highly diverse clinical courses following transplantation, and was caused by clonal ST258 strains with different ompK36 genotypes.
Keywords: Bacteremia; Klebsiella pneumoniae; Klebsiella pneumoniae carbapenemase; ST258; transplantation.
© Copyright 2013 The American Society of Transplantation and the American Society of Transplant Surgeons.
Conflict of interest statement
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Figures
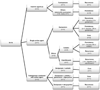
Figure 1. Summary of treatment and outcomes of transplant recipients with CR-Kp bacteremia
* *Patients who died of septic shock within 6 days were excluded because they either received an inactive agent (piperacillin-tazobactam; P2) or received a regimen with a single active agent for only 1 day (P1 and P3). CR-Kp, carbapenem-resistant Klebsiella pneumoniae; n, number; (I), intermediate; (R), resistant.
Figure 2. Distribution of doripenem MICs and ompK36 variants
Doripenem was chosen to represent the carbapenem class since it is the formulary agent at our center. MICs, minimum inhibitory concentrations.
Figure 3. PFGE of representative KPC-Kp isolates from the 17 patients with bacteremia
PFGE dendrogramshowing the relatedness of 41 KPC-producing K. pneumoniae isolates. All KPC-2-producing ST258 strains were clustered in one group, sharing >86.5% similarities. KPC-3-producing ST37 strains were distinct from ST258 strains (<65% similarities). The numbers following the abbreviations for the patients’ samples correspond to days before (number preceded by a minus sign) or after the first positive blood culture (number alone). For example, U-90 denotes urine culture obtained 90 days before the first positive blood culture. B0 denotes the first positive blood culture. T24 denotes tissue culture obtained 24 days after the first positive blood culture. PFGE, pulse-field gel electrophoresis; KPC-K. pneumoniae (KPC-Kp).
Figure 4. Serial ST258, KPC-2-producing strains with ompK36 variants isolated from an intestinal transplant recipient with persistent bacteremia (P9)
Blue and red diamonds represent bloodstream and non-bloodstream strains, respectively.
Similar articles
- Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae.
Landman D, Bratu S, Quale J. Landman D, et al. J Med Microbiol. 2009 Oct;58(Pt 10):1303-1308. doi: 10.1099/jmm.0.012575-0. Epub 2009 Jun 25. J Med Microbiol. 2009. PMID: 19556371 Free PMC article. - KPC-Producing Klebsiella pneumoniae Isolates in Croatia: A Nationwide Survey.
Jelic M, Butic I, Plecko V, Cipris I, Jajic I, Bejuk D, Koscak I, Marinkovic S, Pal MP, Andrasevic AT. Jelic M, et al. Microb Drug Resist. 2016 Dec;22(8):662-667. doi: 10.1089/mdr.2015.0150. Epub 2015 Dec 28. Microb Drug Resist. 2016. PMID: 26709956 - Characterization of the Genetic Background of KPC-2-Producing Klebsiella pneumoniae with Insertion Elements Disrupting the ompK36 Porin Gene.
Wu LT, Guo MK, Ke SC, Lin YP, Pang YC, Nguyen HV, Chen CM. Wu LT, et al. Microb Drug Resist. 2020 Sep;26(9):1050-1057. doi: 10.1089/mdr.2019.0410. Epub 2020 Apr 13. Microb Drug Resist. 2020. PMID: 32283046 - Cluster of bloodstream infections caused by KPC-2 carbapenemase-producing Klebsiella pneumoniae in Manhattan.
Nadkarni AS, Schliep T, Khan L, Zeana CB. Nadkarni AS, et al. Am J Infect Control. 2009 Mar;37(2):121-6. doi: 10.1016/j.ajic.2007.10.013. Am J Infect Control. 2009. PMID: 19249638 - Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece.
Karampatakis T, Antachopoulos C, Iosifidis E, Tsakris A, Roilides E. Karampatakis T, et al. Future Microbiol. 2016 Jun;11:809-23. doi: 10.2217/fmb-2016-0042. Epub 2016 May 20. Future Microbiol. 2016. PMID: 27206024 Review.
Cited by
- Control of infectious mortality due to carbapenemase-producing Klebsiella pneumoniae in hematopoietic stem cell transplantation.
Forcina A, Baldan R, Marasco V, Cichero P, Bondanza A, Noviello M, Piemontese S, Soliman C, Greco R, Lorentino F, Giglio F, Messina C, Carrabba M, Bernardi M, Peccatori J, Moro M, Biancardi A, Nizzero P, Scarpellini P, Cirillo DM, Mancini N, Corti C, Clementi M, Ciceri F. Forcina A, et al. Bone Marrow Transplant. 2017 Jan;52(1):114-119. doi: 10.1038/bmt.2016.234. Epub 2016 Sep 26. Bone Marrow Transplant. 2017. PMID: 27668762 - Risk factors and outcome of infections with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in kidney transplant recipients.
Freire MP, Abdala E, Moura ML, de Paula FJ, Spadão F, Caiaffa-Filho HH, David-Neto E, Nahas WC, Pierrotti LC. Freire MP, et al. Infection. 2015 Jun;43(3):315-23. doi: 10.1007/s15010-015-0743-4. Epub 2015 Feb 18. Infection. 2015. PMID: 25690848 - Molecular epidemiology of antimicrobial resistant microorganisms in the 21th century: a review of the literature.
Genovese C, La Fauci V, D'Amato S, Squeri A, Anzalone C, Costa GB, Fedele F, Squeri R. Genovese C, et al. Acta Biomed. 2020 May 11;91(2):256-273. doi: 10.23750/abm.v91i2.9176. Acta Biomed. 2020. PMID: 32420962 Free PMC article. Review. - Within-Host Genotypic and Phenotypic Diversity of Contemporaneous Carbapenem-Resistant Klebsiella pneumoniae from Blood Cultures of Patients with Bacteremia.
Cheng S, Fleres G, Chen L, Liu G, Hao B, Newbrough A, Driscoll E, Shields RK, Squires KM, Chu TY, Kreiswirth BN, Nguyen MH, Clancy CJ. Cheng S, et al. mBio. 2022 Dec 20;13(6):e0290622. doi: 10.1128/mbio.02906-22. Epub 2022 Nov 29. mBio. 2022. PMID: 36445082 Free PMC article. - Carbapenem-Resistant Klebsiella pneumoniae influences the outcome of early infections in liver transplant recipients.
Barchiesi F, Montalti R, Castelli P, Nicolini D, Staffolani S, Mocchegiani F, Fiorentini A, Manso E, Vivarelli M. Barchiesi F, et al. BMC Infect Dis. 2016 Oct 4;16(1):538. doi: 10.1186/s12879-016-1876-5. BMC Infect Dis. 2016. PMID: 27716164 Free PMC article.
References
- Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: No ESKAPE@! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. - PubMed
- Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63:659–667. - PubMed
- Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. - PubMed
- Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000005/TR/NCATS NIH HHS/United States
- KL2RR024154/RR/NCRR NIH HHS/United States
- 1R01AI090155/AI/NIAID NIH HHS/United States
- R01 AI090155/AI/NIAID NIH HHS/United States
- KL2 RR024154/RR/NCRR NIH HHS/United States
- KL2 TR000146/TR/NCATS NIH HHS/United States
- KL2TR000146/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical