A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia - PubMed (original) (raw)
. 2013 Oct;45(10):1226-1231.
doi: 10.1038/ng.2754. Epub 2013 Sep 8.
Kasmintan A Schrader # 1 2, Esmé Waanders # 3 4, Andrew E Timms # 5, Joseph Vijai # 1 2, Cornelius Miething # 1, Jeremy Wechsler 5, Jun Yang 6, James Hayes 1, Robert J Klein 1 2, Jinghui Zhang 7, Lei Wei 7 3, Gang Wu 7, Michael Rusch 7, Panduka Nagahawatte 7, Jing Ma 8, Shann-Ching Chen 3, Guangchun Song 3, Jinjun Cheng 3 8, Paul Meyers 9, Deepa Bhojwani 10, Suresh Jhanwar 11, Peter Maslak 12, Martin Fleisher 13, Jason Littman 2, Lily Offit 2, Rohini Rau-Murthy 2, Megan Harlan Fleischut 2, Marina Corines 2, Rajmohan Murali 11, Xiaoni Gao 1, Christopher Manschreck 2, Thomas Kitzing 1, Vundavalli V Murty 14, Susana Raimondi 3, Roland P Kuiper 4, Annet Simons 4, Joshua D Schiffman 15, Kenan Onel 16, Sharon E Plon 17, David Wheeler 17, Deborah Ritter 17, David S Ziegler 18, Kathy Tucker 19, Rosemary Sutton 20, Georgia Chenevix-Trench 21, Jun Li 21, David G Huntsman 22, Samantha Hansford 22, Janine Senz 22, Thomas Walsh 23, Ming Lee 23, Christopher N Hahn 24, Kathryn Roberts 3, Mary-Claire King 23, Sarah M Lo 25, Ross L Levine 26, Agnes Viale 27, Nicholas D Socci 28, Katherine L Nathanson 29, Hamish S Scott 24, Mark Daly 30, Steven M Lipkin 31, Scott W Lowe 1, James R Downing 3, David Altshuler 30, John T Sandlund # 10, Marshall S Horwitz # 5, Charles G Mullighan # 3, Kenneth Offit # 1 2
Affiliations
- PMID: 24013638
- PMCID: PMC3919799
- DOI: 10.1038/ng.2754
A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia
Sohela Shah et al. Nat Genet. 2013 Oct.
Abstract
Somatic alterations of the lymphoid transcription factor gene PAX5 (also known as BSAP) are a hallmark of B cell precursor acute lymphoblastic leukemia (B-ALL), but inherited mutations of PAX5 have not previously been described. Here we report a new heterozygous germline variant, c.547G>A (p.Gly183Ser), affecting the octapeptide domain of PAX5 that was found to segregate with disease in two unrelated kindreds with autosomal dominant B-ALL. Leukemic cells from all affected individuals in both families exhibited 9p deletion, with loss of heterozygosity and retention of the mutant PAX5 allele at 9p13. Two additional sporadic ALL cases with 9p loss harbored somatic PAX5 substitutions affecting Gly183. Functional and gene expression analysis of the PAX5 mutation demonstrated that it had significantly reduced transcriptional activity. These data extend the role of PAX5 alterations in the pathogenesis of pre-B cell ALL and implicate PAX5 in a new syndrome of susceptibility to pre-B cell neoplasia.
Figures
Figure 1. Familial Pre-B cell ALL associated with i(9)(q10) and dic(9;v) in two families harboring a novel, recurrent germline p.Gly183Ser variant
a. Family 1 of Puerto Rican ancestry. The proband is denoted by an arrow. Exome sequencing was undertaken in germline DNA from all available affected (IV1, IV5, IV6, III5) and unaffected (IV9, III3, III4) individuals as well as the diagnostic leukemic sample from IV6. p.Gly183Ser variant status denoted by (+/−). b. Family 2 of African-American ancestry. The proband is denoted by an arrow. Exome sequencing was undertaken in diagnostic, remission and relapse leukemic samples from individuals III4, IV1, and IV2. p.Gly183Ser variant status denoted by (+/−). c. Chromosome 9 copy number heat map for SNP6.0 microarray data of germline and tumor samples from three members of Family 2. These data demonstrate the common feature of loss of 9p in the tumor specimens. Note the focal dark blue band denoting homozygous loss of CDKN2A/B in all samples. Blue indicates deletions and red indicates gains. G, germline; D, diagnosis; R, relapse sample. d. The haplotype flanking the p.Gly183Ser mutation. A five SNP haplotype rs7850825 to rs7020413 (Chr9:36.997-37.002 Mb) proximal to the mutation was concordant in both family 1 and family 2. However, the distal end flanking the mutation rs6476606 was discordant.
Figure 2. Recurrent PAX5 mutations in ALL
a. Gene schematic of PAX5 showing the exons (upper grey numbers), amino acid residues (lower grey numbers), protein domains (as denoted by colored legend) and position of the germline p.Gly183Ser variant (in red) in relation to the somatic PAX5 mutations described in this study (n=13, arrows) and somatic mutations described previously in B-ALL,,. Primary leukemic samples with confirmed retention of the germline p.Gly183Ser variant denoted by the square shape (Family 1) and diamond shape (Family 2). In one case of i(9)/dic(9) ALL, we found both a heterozygous Val26Gly and a heterozygous Gln350fs mutation, indicating polyclonality of the tumor. b. Conservation of the octapeptide domain in selected PAX family members.
Figure 3. Attenuated transcriptional activity of PAX5 p.Gly183Ser
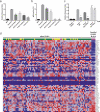
a. Transcriptional activity of PAX5 variants compared to wild-type using a Pax5-dependent reporter gene assay in 293T cells. Bars show mean (± s.e.m.) luciferase activity of six individual experiments with triplicate measurements (PAX5 p.Gly183Val and PAX5 d2-6 four experiments with triplicate measurements). Asterisks indicate significant difference calculated by Dunnett's test (P<0.0001). MIR, MSCV-IRES-mRFP empty vector. b. Transcriptional activity of PAX5 variants using CD79A-dependent sIgM expression in the murine J558LμM plastocytoma cell line. Percentages indicate proportion of mRFP positive cells that show sIgM expression. Bars show mean (± s.e.m.) sIgM expression in two individual experiments with triplicate replicates each. Asterisks indicate significant difference calculated by Dunnett's test (P<0.0001). MIR, MSCV-IRES-mRFP empty vector. c. _PAX5_-dependent reporter gene assay of PAX5 wild-type and PAX5 p.Gly183Ser run in triplicate as above, with or without co-transfection of 0.05μg of Grg4 as indicated. A PAX5 p.Tyr179Glu (Y179E) mutant that is deficient in binding to Grg4 and empty vector were used as controls. Asterisks indicate significant differences by two-tailed t-test (p<0.0001). d. Heatmap of PAX5 activated genes in mature B cells. Four samples from family 2 (diagnosis and relapse samples from individuals IV1 and IV2) show differential expression of PAX5 activated genes when compared to a group of 139 sporadic B-ALL cases. This indicates an effect of the pGly183Ser mutation on PAX5 function. Red indicates high expression, blue represents low expression. PAX5 mutation status is indicated by the colors above the samples. Green indicates wild type PAX5, yellow indicates heterozygosity for a PAX5 mutation, and pink indicates biallelic PAX5 mutations.
Comment in
- Germline PAX5 mutations and B cell leukemia.
Hyde RK, Liu PP. Hyde RK, et al. Nat Genet. 2013 Oct;45(10):1104-5. doi: 10.1038/ng.2778. Nat Genet. 2013. PMID: 24071841
Similar articles
- Germline PAX5 mutations and B cell leukemia.
Hyde RK, Liu PP. Hyde RK, et al. Nat Genet. 2013 Oct;45(10):1104-5. doi: 10.1038/ng.2778. Nat Genet. 2013. PMID: 24071841 - Germline PAX5 mutation predisposes to familial B-cell precursor acute lymphoblastic leukemia.
Duployez N, Jamrog LA, Fregona V, Hamelle C, Fenwarth L, Lejeune S, Helevaut N, Geffroy S, Caillault A, Marceau-Renaut A, Poulain S, Roche-Lestienne C, Largeaud L, Prade N, Dufrechou S, Hébrard S, Berthon C, Nelken B, Fernandes J, Villenet C, Figeac M, Gerby B, Delabesse E, Preudhomme C, Broccardo C. Duployez N, et al. Blood. 2021 Mar 11;137(10):1424-1428. doi: 10.1182/blood.2020005756. Blood. 2021. PMID: 33036026 No abstract available. - PAX5 biallelic genomic alterations define a novel subgroup of B-cell precursor acute lymphoblastic leukemia.
Bastian L, Schroeder MP, Eckert C, Schlee C, Tanchez JO, Kämpf S, Wagner DL, Schulze V, Isaakidis K, Lázaro-Navarro J, Hänzelmann S, James AR, Ekici A, Burmeister T, Schwartz S, Schrappe M, Horstmann M, Vosberg S, Krebs S, Blum H, Hecht J, Greif PA, Rieger MA, Brüggemann M, Gökbuget N, Neumann M, Baldus CD. Bastian L, et al. Leukemia. 2019 Aug;33(8):1895-1909. doi: 10.1038/s41375-019-0430-z. Epub 2019 Mar 6. Leukemia. 2019. PMID: 30842609 - Functional damaging germline variants in ETV6, IKZF1, PAX5 and RUNX1 predisposing to B-cell precursor acute lymphoblastic leukemia.
Wagener R, Elitzur S, Brozou T, Borkhardt A. Wagener R, et al. Eur J Med Genet. 2023 Apr;66(4):104725. doi: 10.1016/j.ejmg.2023.104725. Epub 2023 Feb 9. Eur J Med Genet. 2023. PMID: 36764385 Review. - PAX5 fusion genes in acute lymphoblastic leukemia: A literature review.
Fouad FM, Eid JI. Fouad FM, et al. Medicine (Baltimore). 2023 May 19;102(20):e33836. doi: 10.1097/MD.0000000000033836. Medicine (Baltimore). 2023. PMID: 37335685 Free PMC article. Review.
Cited by
- Mycovirus-Containing Aspergillus flavus Alters Transcription Factors in Normal and Acute Lymphoblastic Leukemia Cells.
Tebbi CK, Yan J, Sahakian E, Mediavilla-Varela M, Pinilla-Ibarz J, Patel S, Rottinghaus GE, Liu RY, Dennison C. Tebbi CK, et al. Int J Mol Sci. 2024 Sep 26;25(19):10361. doi: 10.3390/ijms251910361. Int J Mol Sci. 2024. PMID: 39408690 Free PMC article. - Diverse mechanisms of leukemogenesis associated with PAX5 germline mutation.
Bettini LR, Fazio G, Saitta C, Piazza R, Palamini S, Buracchi C, Rebellato S, Santoro N, Simone C, Biondi A, Cazzaniga G. Bettini LR, et al. Leukemia. 2024 Nov;38(11):2479-2482. doi: 10.1038/s41375-024-02399-0. Epub 2024 Sep 10. Leukemia. 2024. PMID: 39256601 Free PMC article. No abstract available. - Unraveling Copy Number Alterations in Pediatric B-Cell Acute Lymphoblastic Leukemia: Correlation with Induction Phase Remission Using MLPA.
Aisyi M, Andriastuti M, Kosasih AS, Utomo ARH, Saputra F, Tjitra Sari T, Sjakti HA, Dwijayanti F, Harimurti K, Gatot D. Aisyi M, et al. Asian Pac J Cancer Prev. 2024 Jul 1;25(7):2421-2426. doi: 10.31557/APJCP.2024.25.7.2421. Asian Pac J Cancer Prev. 2024. PMID: 39068576 Free PMC article. - SJPedPanel: A Pan-Cancer Gene Panel for Childhood Malignancies to Enhance Cancer Monitoring and Early Detection.
Kolekar P, Balagopal V, Dong L, Liu Y, Foy S, Tran Q, Mulder H, Huskey ALW, Plyler E, Liang Z, Ma J, Nakitandwe J, Gu J, Namwanje M, Maciaszek J, Payne-Turner D, Mallampati S, Wang L, Easton J, Klco JM, Ma X. Kolekar P, et al. Clin Cancer Res. 2024 Sep 13;30(18):4100-4114. doi: 10.1158/1078-0432.CCR-24-1063. Clin Cancer Res. 2024. PMID: 39047169 Free PMC article. - The Diverse Roles of ETV6 Alterations in B-Lymphoblastic Leukemia and Other Hematopoietic Cancers.
Monovich AC, Gurumurthy A, Ryan RJH. Monovich AC, et al. Adv Exp Med Biol. 2024;1459:291-320. doi: 10.1007/978-3-031-62731-6_13. Adv Exp Med Biol. 2024. PMID: 39017849 Review.
References
- Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. - PubMed
- Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–66. - PubMed
- Hemminki K, Jiang Y. Risks among siblings and twins for childhood acute lymphoid leukaemia: results from the Swedish Family-Cancer Database. Leukemia. 2002;16:297–8. - PubMed
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01DK58161/DK/NIDDK NIH HHS/United States
- K12 GM084897/GM/NIGMS NIH HHS/United States
- T32 GM007454/GM/NIGMS NIH HHS/United States
- CAPMC/ CIHR/Canada
- P30 CA021765/CA/NCI NIH HHS/United States
- R01 DK058161/DK/NIDDK NIH HHS/United States
- R01 CA138836/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- T32GM007454/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases