The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair - PubMed (original) (raw)
The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair
Alexander J Garvin et al. EMBO Rep. 2013 Nov.
Abstract
SUMO conjugation is known to occur in response to double-stranded DNA breaks in mammalian cells, but whether SUMO deconjugation has a role remains unclear. Here, we show that the SUMO/Sentrin/Smt3-specific peptidase, SENP7, interacts with the chromatin repressive KRAB-associated protein 1 (KAP1) through heterochromatin protein 1 alpha (HP1α). SENP7 promotes the removal of SUMO2/3 from KAP1 and regulates the interaction of the chromatin remodeler CHD3 with chromatin. Consequently, in the presence of CHD3, SENP7 is required for chromatin relaxation in response to DNA damage, for homologous recombination repair and for cellular resistance to DNA-damaging agents. Thus, deSUMOylation by SENP7 is required to promote a permissive chromatin environment for DNA repair.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
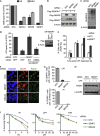
The SENP7 protease is required for HR repair. (A) Chain-editing SUMO proteases SENP6 and SENP7 are required for DSB repair. HeLa DR3-GFP (HR repair) and EJ5-GFP (NHEJ) reporters were treated with non-targeting control, BRCA1, SENP6 or SENP7 siRNA before transfection with Sce-I. (All bars throughout show standard error about the mean of three independent experiments). (B) Knockdown efficiency of SENP6/7. HEK293 expressing Flag-SENP6/SENP7 and treated with indicated siRNA. (C) Depletion of SENP6 and SENP7 increases SUMO2/3 conjugates. HEK293 transfected with indicated siRNA and lysates probed with SUMO2/3 antibody and anti-β-actin. (D) siRNA-resistant WT-SENP7, but not catalytic mutant, C992A, can compensate for SENP7 depletion in HR repair assays. HeLa DR3 depleted of endogenous SENP7 before transfection with Sce-I, RFP and siRNA-resistant Flag-SENP7. % HR repair relative to non-targeting control is shown. Inset shows WB of Flag-SENP7 expression and β-actin loading control. (E) Clearance of γH2AX foci after CPT treatment. U2OS depleted of SENP7 and treated with 1μM CPT (1 h), before washing and recovery. Cells stained against γH2AX. % cells with >5 foci were scored. (F–H) RAD51 foci are reduced in SENP7-depleted cells. U2OS transfected with siRNA before 2.5 Gy IR or 1 μM CPT/1 h. Cells were stained 4 h later for RAD51 and CENPF to identify G2-phase cells. % cells with >5 foci scored in CENPF-positive cells (G). Representative images of IR-treated cells are shown in (F). Immunoblot from parallel experiment lysed and blotted with RAD51 (H). (I) SENP7 depletion sensitizes cells to IR, CPT and PARP inhibition (4AN). Clonogenic assay of HeLa treated with indicated siRNA followed by doses of 4AN, IR or CPT. 4AN, 4-amino-1-8 naphthalimide; CPT, camptothecin; DSB, double-strand break repair; HR, homologous recombination; IR, ionizing radiation; NTC, non-targeting control; PARP, poly ADP ribose polymerase; siRNA, small interfering RNA; UT, untreated; WB, western blot; WT, wild type.
Figure 2
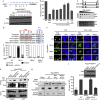
SENP7 SIMs and HP1-box are required to promote HR repair. (A) SIM mutants of SENP7. Location of each SIM (red lines) and the catalytic domain (yellow oval) are shown. Below, shows expression of Flag-SENP7 SIM mutants in HeLa. (B) SENP7 SIMs required for HR activity. HR rescue repair assays using Flag-siR-SIM mutants. % repair is measured relative to WT-SENP7. (C) SENP7 SIM 6 and 7 bind to polySUMO2. Cartoon of WT and SIM6 and 7 mutant fragments (aa 549–655). Spotted fragments were stained with Ponceau, incubated with polySUMO2, washed and probed for bound SUMO2. (D) SENP7 contains a NES adjacent to its HP1-box. NES and HP1-box consensus with the SENP7 NES and HP1 sequence and mutants made in this study. (E) Nuclear export of HP1-box mutant. U2OS expressing Flag-SENP7-WT and indicated mutants. 1 h before fixation and Flag staining, cells were treated with 10 μM cycloheximide and 6 nM Leptomycin-B or DMSO. Scale bar, 10 μm (F) Quantification of SENP7 localization from experiments described in (E) scored in 100 cells in three independent experiments (mean shown). (G) The NESm-HP1m mutant of SENP7 does not interact with endogenous HP1α. HEK293 expressing Flag-SENP7 constructs were untreated or exposed to CPT, lysed and immunoprecipitated with Flag. Eluates were blotted with Flag and HP1α. Lower panels=5% inputs. (H) SENP7 NESm-HP1m mutant is not localized with chromatin. HEK293 lysates expressing Flag-SENP7 constructs, separated into soluble and insoluble (pellet) fractions. Lysates were probed with Flag (SENP7), H2A (chromatin marker) and GAPDH (soluble marker). (I) HP1-box is required for HR repair. HR repair assays performed as for Fig 1C. % repair is shown relative to WT-SENP7. Inset image shows expression of mutants. CPT, camptothecin; DMSO, dimethyl sulphoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HP1α, heterochromatin protein 1 alpha; HR, homologous recombination; NES, nuclear export sequence; WT, wild type.
Figure 3
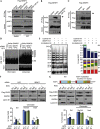
SENP7 deSUMOylates KAP1 and regulates CHD3–chromatin association. (A) SENP7 interacts with KAP1. HEK293 expressing Flag-SENP7 was mock or CPT treated, lysed, immunoprecipitated and immunoblotted with Flag and KAP1. pSer824-KAP1 used as a marker of DNA damage. (B) SENP7–C992A interaction with KAP1 requires the HP1-box. HEK293 expressing Flag-C992A-SENP7 or Flag-C992A-NESm-HP1m-SENP7 immunoprecipitated and immunoblotted with Flag and KAP1. (C) HP1α interaction with WT and SENP7-C992A. HEK293 expressing Flag-WT and Flag-C992A-SENP7 were immunoprecipitated as above and blotted for HP1α. (D) KAP1 SUMO2ylation is increased on SENP7 depletion. 6xHis-SUMO2 expressing HeLa depleted with NTC or SENP7 siRNAs before CPT, lysed in 8 M urea and incubated with nickel-agarose. Eluates and 5% input were immunoblotted with KAP1. S=number of His-SUMO2 modifications.(E) DeSUMOylation of KAP1-polySUMO in vitro. Auto-SUMO2ylated GST-KAP1 incubated with full-length in vitro translated SENP7 for 1–3 h. Beads were washed, boiled and KAP1-SUMOylation detected on SPYRO ruby stained 6% gels. S0=unSUMOylated GST-KAP1. Quantification of SUMOylated KAP1, each SUMOylated species is calculated as % of total KAP1/lane. (F) SENP7 depletion increases CHD3–chromatin retention. HEK293 expressing WT Flag-CHD3 treated with indicated siRNA and NCS (200 ng/ml 1 h). Lysates were fractionated into soluble (Sol) and chromatin. The chromatin was subsequently fractionated into salt-soluble (NaS) and pellet (Ins) sub-fractions. Lysates were blotted with Flag and fraction markers. The proportions of Flag-CHD3 were quantified from three experiments below. Significance was assessed using Student’s _t_-test; *<0.05. (G) KAP1-4xSUMO2 increases CHD3 chromatin retention. HEK293 transfected with Flag-CHD3 and KAP1 or KAP1-4xSUMO2 (illustrated) and treated as for F. Quantification was as for F. Significance was assessed using Student’s _t_-test; *<0.05. CPT, camptothecin; GST, glutathione _S_-transferase; HP1α, heterochromatin protein 1 alpha; KAP1, KRAB-associated protein 1; NCS, neocarzinostatin; NTC, non-targeting control; siRNA, small interfering RNA; WT, wild type.
Figure 4
SENP7 regulates chromatin decondensation required for HR repair. (A) SENP7 is required for chromatin relaxation in response to DNA damage. HEK293 transfected with siRNA before treatment with NCS. Nuclei were incubated with MNase (0.25 U/5 min). Data are represented as % signal/lane determined by densitometry. (B) SENP7 is required for localized euchromatin relaxation. Cells stably expressing oestrogen receptor-fused IPpoI transfected with siRNA and treated with vehicle or 4-OHT to induce DSB’s. Euchromatin accessible to digestion was assayed using EpiQ assay. Chromatin was digested and PCR amplification of the chromosome 1 site (DAB1 gene intron) adjacent to the IPpoI endonuclease target site was undertaken. Graph shows Qc (amplification efficiency) of (digested/undigested) for vehicle/4-OHT treatment. (C) CHD3 depletion restores HR repair in SENP7-depleted cells. HR repair assay performed as for Fig 1C. % GFP/RFP (HR repair) relative to GFP/RFP in NTC is shown. (D) CHD3 depletion confers PARP-inhibitor resistance of SENP7-depleted cells. Clonogenic assay of HeLa transfected with indicated siRNA followed by treatment with 4AN. (E) CHD3 depletion restores chromatin decondensation in SENP7-depleted cells. Cells transfected with indicated siRNAs before NCS treatment and MNase assay as in Fig 4A. (F) γH2AX clearance is restored by depletion of Chd3 in Senp7-depleted mouse cells. NIH3T3 transfected with NTC or Senp7 siRNA, exposed to 2.5 Gy IR and allowed to recover for indicated times before staining with antibody to γH2AX. Cells with >5 foci were counted (100 cells, in three independent experiments). (G). Model of SENP7 in promoting chromatin state in the presence and absence of DNA damage. (i) SENP7 and KAP1 are co-located on chromatin. SENP7 constitutively deSUMOylates KAP1 preventing formation of polySUMO2/3-KAP1. SUMO-KAP1 associates with NuRD subunit CHD3 and histone methyltransferase SETDB1 via SUMO interacting motifs. (ii) KAP1-SUMO homeostasis is a two-step process by which SENP7 deconjugates the polySUMO2/3 KAP1 and other SUMO proteases, such as SENP1 deconjugate the mono-SUMO2/3 KAP1. (iii) Following induction of DNA damage response, KAP1 is phosphorylated at Ser824 by ATM. KAP1 phosphorylation interferes with CHD3SIM–SUMO-KAP1 interaction, allowing dispersion of CHD3–NuRD complexes resulting in chromatin relaxation and repair of DNA. (iv) In the absence of SENP7, KAP1 is hyperSUMO2ylated. The increased SUMO2 conjugates in the vicinity of CHD3SIM negate the interference provided by the negatively charged KAP1-pSer824 and CHD3–NuRD is not released from chromatin and remodelling does not occur. (vi) Potentially prolonged absence of SENP7 promotes hyperSUMOylation of KAP1 leading to excessive remodeller accumulation and (through SETDB1) increased methylation of H3K9. This results in spread of heterochromatin factors and condensed chromatin. 4AN, 4-amino-1-8 naphthalimide; DSB, double-strand break repair; GFP, green fluorescence protein; HR, homologous recombination; IR, ionizing radiation; KAP1, KRAB-associated protein 1; NCS, neocarzinostatin; NTC, non-targeting control; NuRD, nucleosome remodelling and deacetylation; PARP, poly ADP ribose polymerase; PCR, polymerase chain reaction; RFP, red fluorescent protein; SETDB1, SET domain, bifurcated 1; siRNA, small interfering RNA.
Similar articles
- The SUMO protease SENP1 and the chromatin remodeler CHD3 interact and jointly affect chromatin accessibility and gene expression.
Rodríguez-Castañeda F, Lemma RB, Cuervo I, Bengtsen M, Moen LM, Ledsaak M, Eskeland R, Gabrielsen OS. Rodríguez-Castañeda F, et al. J Biol Chem. 2018 Oct 5;293(40):15439-15454. doi: 10.1074/jbc.RA118.002844. Epub 2018 Aug 6. J Biol Chem. 2018. PMID: 30082317 Free PMC article. - Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair.
Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. Klement K, et al. J Cell Biol. 2014 Dec 22;207(6):717-33. doi: 10.1083/jcb.201405077. J Cell Biol. 2014. PMID: 25533843 Free PMC article. - KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response.
Goodarzi AA, Kurka T, Jeggo PA. Goodarzi AA, et al. Nat Struct Mol Biol. 2011 Jun 5;18(7):831-9. doi: 10.1038/nsmb.2077. Nat Struct Mol Biol. 2011. PMID: 21642969 - Chromatin modification and NBS1: their relationship in DNA double-strand break repair.
Saito Y, Zhou H, Kobayashi J. Saito Y, et al. Genes Genet Syst. 2016;90(4):195-208. doi: 10.1266/ggs.15-00010. Epub 2015 Nov 25. Genes Genet Syst. 2016. PMID: 26616756 Review. - Chromatin structure in double strand break repair.
Gospodinov A, Herceg Z. Gospodinov A, et al. DNA Repair (Amst). 2013 Oct;12(10):800-10. doi: 10.1016/j.dnarep.2013.07.006. Epub 2013 Aug 6. DNA Repair (Amst). 2013. PMID: 23919923 Review.
Cited by
- Moving Mountains-The BRCA1 Promotion of DNA Resection.
Densham RM, Morris JR. Densham RM, et al. Front Mol Biosci. 2019 Sep 3;6:79. doi: 10.3389/fmolb.2019.00079. eCollection 2019. Front Mol Biosci. 2019. PMID: 31552267 Free PMC article. Review. - The Chromatin Structure of CRISPR-Cas9 Target DNA Controls the Balance between Mutagenic and Homology-Directed Gene-Editing Events.
Janssen JM, Chen X, Liu J, Gonçalves MAFV. Janssen JM, et al. Mol Ther Nucleic Acids. 2019 Jun 7;16:141-154. doi: 10.1016/j.omtn.2019.02.009. Epub 2019 Feb 20. Mol Ther Nucleic Acids. 2019. PMID: 30884291 Free PMC article. - In the moonlight: non-catalytic functions of ubiquitin and ubiquitin-like proteases.
Campos Alonso M, Knobeloch KP. Campos Alonso M, et al. Front Mol Biosci. 2024 Feb 22;11:1349509. doi: 10.3389/fmolb.2024.1349509. eCollection 2024. Front Mol Biosci. 2024. PMID: 38455765 Free PMC article. Review. - Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway.
Li J, Lu D, Dou H, Liu H, Weaver K, Wang W, Li J, Yeh ETH, Williams BO, Zheng L, Yang T. Li J, et al. Nat Commun. 2018 Jan 10;9(1):143. doi: 10.1038/s41467-017-02413-3. Nat Commun. 2018. PMID: 29321472 Free PMC article. - SPOP E3 Ubiquitin Ligase Adaptor Promotes Cellular Senescence by Degrading the SENP7 deSUMOylase.
Zhu H, Ren S, Bitler BG, Aird KM, Tu Z, Skordalakes E, Zhu Y, Yan J, Sun Y, Zhang R. Zhu H, et al. Cell Rep. 2015 Nov 10;13(6):1183-1193. doi: 10.1016/j.celrep.2015.09.083. Epub 2015 Oct 29. Cell Rep. 2015. PMID: 26527005 Free PMC article.
References
- Psakhye I & Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820 - PubMed
- Bekker-Jensen S & Mailand N (2011) The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett 585: 2914–2919 - PubMed
- Ziv Y Bielopolski D Galanty Y Lukas C Taya Y Schultz DC Lukas J Bekker-Jensen S Bartek J & Shiloh Y (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous