GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia - PubMed (original) (raw)
. 2013 Dec 5;122(24):3908-17.
doi: 10.1182/blood-2013-07-515148. Epub 2013 Sep 10.
Kate Alford, Georgina Hall, Gaetan Juban, Helen Richmond, Alice Norton, Grant Vallance, Kelly Perkins, Emanuele Marchi, Simon McGowan, Anindita Roy, Gillian Cowan, Mark Anthony, Amit Gupta, John Ho, Sabita Uthaya, Anna Curley, Shree Vishna Rasiah, Timothy Watts, Richard Nicholl, Alison Bedford-Russell, Raoul Blumberg, Angela Thomas, Brenda Gibson, Chris Halsey, Pek-Wan Lee, Sunit Godambe, Connor Sweeney, Neha Bhatnagar, Anne Goriely, Peter Campbell, Paresh Vyas; Oxford-Imperial Down Syndrome Cohort Study Group
Affiliations
- PMID: 24021668
- PMCID: PMC3995281
- DOI: 10.1182/blood-2013-07-515148
GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia
Irene Roberts et al. Blood. 2013.
Abstract
Transient abnormal myelopoiesis (TAM), a preleukemic disorder unique to neonates with Down syndrome (DS), may transform to childhood acute myeloid leukemia (ML-DS). Acquired GATA1 mutations are present in both TAM and ML-DS. Current definitions of TAM specify neither the percentage of blasts nor the role of GATA1 mutation analysis. To define TAM, we prospectively analyzed clinical findings, blood counts and smears, and GATA1 mutation status in 200 DS neonates. All DS neonates had multiple blood count and smear abnormalities. Surprisingly, 195 of 200 (97.5%) had circulating blasts. GATA1 mutations were detected by Sanger sequencing/denaturing high performance liquid chromatography (Ss/DHPLC) in 17 of 200 (8.5%), all with blasts >10%. Furthermore low-abundance GATA1 mutant clones were detected by targeted next-generation resequencing (NGS) in 18 of 88 (20.4%; sensitivity ∼0.3%) DS neonates without Ss/DHPLC-detectable GATA1 mutations. No clinical or hematologic features distinguished these 18 neonates. We suggest the term "silent TAM" for neonates with DS with GATA1 mutations detectable only by NGS. To identify all babies at risk of ML-DS, we suggest GATA1 mutation and blood count and smear analyses should be performed in DS neonates. Ss/DPHLC can be used for initial screening, but where GATA1 mutations are undetectable by Ss/DHPLC, NGS-based methods can identify neonates with small GATA1 mutant clones.
Figures
Figure 1
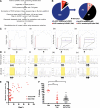
Hematologic abnormalities and GATA1 mutation analysis by Ss/DHPLC in neonates with DS. (A) Percentage of blasts on blood films from the first week of life in 200 neonates with DS, 17 with TAM (red circles) and 183 without TAM (black circles). (B) Photomicrographs of typical blast cells in a neonate with TAM (top) and in a DS neonate without TAM (bottom). (C) GATA1 mutation analysis in TAM by Ss and DHPLC. (Ci,ii) Mutation analysis of sample DST11. The mutation is detected by both Ss and DHPLC. (Ci) Sanger sequence trace. The arrow points the start of a double sequence trace indicative of an acquired GATA1 mutation. (Cii) DHPLC trace from the same sample (red line, mutant; black line, normal). (Ciii,iv) Mutation analysis of sample DST9. The mutation is detected by DHPLC but not by Ss. (Ciii) Sequence trace. (Civ) DHPLC trace from the same sample (red line, mutant; black line, normal). (D,F,H) Scatter graphs of hematocrit (D), platelet counts (F), and leukocytes (H) in 200 DS neonates in the first week of life, 17 with TAM (red circles) and 183 without TAM (black circles). The horizontal lines show the upper and/or lower limits of the normal neonatal laboratory range (see supplemental Methods). (E,G,I) Photomicrographs of erythrocyte (E), platelet (G), and leukocyte (I) morphologic abnormalities in neonates with DS. (E) Top left: macrocytes (black arrowheads); top right: target cells (white arrowheads); bottom left: dyserythropoietic erythroblasts (fine black arrow); bottom right: basophilic stippling (gray arrow). (G) Examples of giant platelets (GP) (black arrowhead) and megakaryoblasts (white arrowhead), megakaryocyte fragments (MK fragment), and circulating megakaryocytes (MKs) in blood films from DS neonates without TAM (top row) and with TAM (bottom row). (I) Top left: hypogranular neutrophil; top right: pseudo-Pelger neutrophil; bottom left: monocyte with stellate nucleus; bottom right, dysplastic basophil. Scale bars indicate 10 μm. WBC, white blood cell.
Figure 2
GATA1 mutation analysis in DS neonates with TAM and silent TAM. (A) Flow diagram of preparation and analysis of samples for deep sequencing. (B) Pie charts of GATA1 mutation analysis of the 200 babies in the cohort by standard Ss/DHPLC (left) and NGS (right). (C) Examples of base-pair plots from NGR analysis of patient samples (mutation indicated by arrows) with (D) corresponding pyrosequencing traces below (mutant peaks indicated by arrows). On the x-axis is the position along the GATA1 exon 2 amplicon (432 base pairs). On the y-axis is the read depth at different positions along the amplicon. Therefore, the black line trace shows the number of reads mapping to GATA1 sequence at different positions along the amplicon. At the position of the black arrowhead, a mutation was introduced into the PCR primer (mapping outside GATA1 exon 2) so that all PCR products would have a unique tag. This introduced mutation is detected by the blue line. All PCR products have this introduced mutation as the height of the blue line is to the level of black trace (total number of mapping reads). Sequence analysis also shows there are 2 common single-nucleotide polymorphism at positions rs62600348 T>G and rs66717003 T>G (indicated by the star) in the amplicon that map to position 48649449 and 48649456 within GATA1 exon 2. Nucleotide 0 is the first nucleotide of GATA1 exon 1 including exons and introns. NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) was used. The location of GATA1 mutation is indicated by the black arrow. (Ci,Di) Patient sample DST11 with a 7 bp duplication at position 48649625 previously detected by Ss/DHPLC. (Cii,Dii) Patient sample DST9 with an insertion of 7 bp at position 48649670 previously detected by DHPLC only. (Ciii,Diii) Patient sample DS158 with a 2 bp deletion at position 48649552 detected by NGS only and confirmed by pyrosequencing. (Civ,Div) Patient sample DS79 with a point mutation at position 48649565 detected by NGS only but not detectable by pyrosequencing. (E) Relationship between GATA1 mutant clone size (y-axis) as determined by NGR and % blasts detected by morphology. (F) Distribution of % blasts in TAM (n = 17) (filled red circles, left), silent TAM (n = 88) (open red cell circles, middle) and in samples without a GATA1 mutation detected by NGR (n = 70) (filled black circles, right).
Figure 3
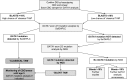
Algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Suggested algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Evaluation of a blood smear and CBC can be used as an immediate screening step to identify DS neonates with “classical” TAM who may require early treatment (especially where GATA1 analysis is unavailable or delayed). As a next step, GATA1 mutation analysis by Ss/DHPLC will quickly identify DS neonates with large mutant GATA1 clones. For DS neonates without mutations detected by Ss/DHPLC, NGS is the most reliable way of identifying low-abundance GATA1 mutations, allowing pediatric hematology follow-up to be limited to those at risk of transformation rather than all DS babies with peripheral blood blasts. Monitoring of all DS children with GATA1 mutations until the age of 5 years is recommended. This can be done using serial CBC/smears with GATA1 mutation analysis as indicated (eg, for persistent cytopenias). For the small number of DS babies with blasts >10% who have no detectable GATA1 mutations by NGS, more detailed studies to exclude the presence of rare GATA1 deletions are suggested.
Comment in
- Neonatal GATA1 mutant clones under the radar.
Taub JW. Taub JW. Blood. 2013 Dec 5;122(24):3851-3. doi: 10.1182/blood-2013-10-527598. Blood. 2013. PMID: 24311713
Similar articles
- A sensitive and inexpensive high-resolution melting-based testing algorithm for diagnosis of transient abnormal myelopoiesis and myeloid leukemia of Down syndrome.
Camargo R, de Castro Moreira Dos Santos A Jr, Cândido Guido B, Lemos Mendanha Cavalcante L, Silva Dias AC, Mendonça de Pontes R, Magalhães Furtado F, Feitosa Salviano C, Tiziani V, Martins Córdoba JC, Quezado Magalhães IM. Camargo R, et al. Pediatr Blood Cancer. 2022 Nov;69(11):e29866. doi: 10.1002/pbc.29866. Epub 2022 Jul 11. Pediatr Blood Cancer. 2022. PMID: 35731576 - Complete blood count differences in a cohort of Down syndrome neonates with transient abnormal myelopoiesis screened for GATA1 pathogenic variants.
Orozco-Vela M, Corona-Rivera A, Cruz-Osorio RM, Mendoza-Maldonado L, Márquez-Mora A, Barba-Barba CC, Peña-Padilla C, Baldomero-López A, Bobadilla-Morales L, Corona-Rivera JR. Orozco-Vela M, et al. Am J Med Genet A. 2020 Sep;182(9):2085-2093. doi: 10.1002/ajmg.a.61748. Epub 2020 Jul 18. Am J Med Genet A. 2020. PMID: 32681702 - Recent advances in the understanding of transient abnormal myelopoiesis in Down syndrome.
Watanabe K. Watanabe K. Pediatr Int. 2019 Mar;61(3):222-229. doi: 10.1111/ped.13776. Epub 2019 Mar 4. Pediatr Int. 2019. PMID: 30593694 Review. - Highly sensitive detection of GATA1 mutations in patients with myeloid leukemia associated with Down syndrome by combining Sanger and targeted next generation sequencing.
Terui K, Toki T, Taga T, Iwamoto S, Miyamura T, Hasegawa D, Moritake H, Hama A, Nakashima K, Kanezaki R, Kudo K, Saito AM, Horibe K, Adachi S, Tomizawa D, Ito E. Terui K, et al. Genes Chromosomes Cancer. 2020 Mar;59(3):160-167. doi: 10.1002/gcc.22816. Epub 2019 Oct 21. Genes Chromosomes Cancer. 2020. PMID: 31606922 - Biology and management of transient abnormal myelopoiesis (TAM) in children with Down syndrome.
Roy A, Roberts I, Vyas P. Roy A, et al. Semin Fetal Neonatal Med. 2012 Aug;17(4):196-201. doi: 10.1016/j.siny.2012.02.010. Epub 2012 Mar 14. Semin Fetal Neonatal Med. 2012. PMID: 22421527 Review.
Cited by
- Down syndrome-associated leukaemias: current evidence and challenges.
Mason NR, Cahill H, Diamond Y, McCleary K, Kotecha RS, Marshall GM, Mateos MK. Mason NR, et al. Ther Adv Hematol. 2024 Jul 23;15:20406207241257901. doi: 10.1177/20406207241257901. eCollection 2024. Ther Adv Hematol. 2024. PMID: 39050114 Free PMC article. Review. - GATA1 in Normal and Pathologic Megakaryopoiesis and Platelet Development.
Takasaki K, Chou ST. Takasaki K, et al. Adv Exp Med Biol. 2024;1459:261-287. doi: 10.1007/978-3-031-62731-6_12. Adv Exp Med Biol. 2024. PMID: 39017848 Review. - Insights into the Clinical, Biological and Therapeutic Impact of Copy Number Alteration in Cancer.
Carey-Smith SL, Kotecha RS, Cheung LC, Malinge S. Carey-Smith SL, et al. Int J Mol Sci. 2024 Jun 21;25(13):6815. doi: 10.3390/ijms25136815. Int J Mol Sci. 2024. PMID: 38999925 Free PMC article. Review. - Aberrant stem cell and developmental programs in pediatric leukemia.
Ling RE, Cross JW, Roy A. Ling RE, et al. Front Cell Dev Biol. 2024 Mar 27;12:1372899. doi: 10.3389/fcell.2024.1372899. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38601080 Free PMC article. Review. - A Trisomy 21-linked Hematopoietic Gene Variant in Microglia Confers Resilience in Human iPSC Models of Alzheimer's Disease.
Jin M, Ma Z, Dang R, Zhang H, Kim R, Xue H, Pascual J, Finkbeiner S, Head E, Liu Y, Jiang P. Jin M, et al. bioRxiv [Preprint]. 2024 Mar 14:2024.03.12.584646. doi: 10.1101/2024.03.12.584646. bioRxiv. 2024. PMID: 38559257 Free PMC article. Preprint.
References
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355(9199):165–169. - PubMed
- Lange BJ, Kobrinsky N, Barnard DR, et al. Distinctive demography, biology, and outcome of acute myeloid leukemia and myelodysplastic syndrome in children with Down syndrome: Children’s Cancer Group Studies 2861 and 2891. Blood. 1998;91(2):608–615. - PubMed
- Ahmed M, Sternberg A, Hall G, et al. Natural history of GATA1 mutations in Down syndrome. Blood. 2004;103(7):2480–2489. - PubMed
- Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;101(11):4301–4304. - PubMed
- Rainis L, Bercovich D, Strehl S, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102(3):981–986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 091182/WT_/Wellcome Trust/United Kingdom
- G1000729/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_U137961146/MRC_/Medical Research Council/United Kingdom
- SCD/08/CSO_/Chief Scientist Office/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical