Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving - PubMed (original) (raw)
Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving
Yuchao Zhang et al. Mol Biol Evol. 2013 Dec.
Erratum in
- Genes that Escape X-Inactivation in Humans Have High Intraspecific Variability in Expression, Are Associated with Mental Impairment but Are Not Slow Evolving.
Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, Hu L, Urrutia AO, Kong X, Hurst LD. Zhang Y, et al. Mol Biol Evol. 2016 Jan;33(1):302. doi: 10.1093/molbev/msv180. Epub 2015 Nov 13. Mol Biol Evol. 2016. PMID: 26568614 Free PMC article. No abstract available.
Abstract
In female mammals most X-linked genes are subject to X-inactivation. However, in humans some X-linked genes escape silencing, these escapees being candidates for the phenotypic aberrations seen in polyX karyotypes. These escape genes have been reported to be under stronger purifying selection than other X-linked genes. Although it is known that escape from X-inactivation is much more common in humans than in mice, systematic assays of escape in humans have to date employed only interspecies somatic cell hybrids. Here we provide the first systematic next-generation sequencing analysis of escape in a human cell line. We analyzed RNA and genotype sequencing data obtained from B lymphocyte cell lines derived from Europeans (CEU) and Yorubans (YRI). By replicated detection of heterozygosis in the transcriptome, we identified 114 escaping genes, including 76 not previously known to be escapees. The newly described escape genes cluster on the X chromosome in the same chromosomal regions as the previously known escapees. There is an excess of escaping genes associated with mental retardation, consistent with this being a common phenotype of polyX phenotypes. We find both differences between populations and between individuals in the propensity to escape. Indeed, we provide the first evidence for there being both hyper- and hypo-escapee females in the human population, consistent with the highly variable phenotypic presentation of polyX karyotypes. Considering also prior data, we reclassify genes as being always, never, and sometimes escape genes. We fail to replicate the prior claim that genes that escape X-inactivation are under stronger purifying selection than others.
Keywords: X-inactivation; expression evolution; rate of evolution.
Figures
Fig. 1.
The location of escape genes in CEU and YRI cluster in similar chromosomal locations. The genes found in more than three individuals and in greater than 50% of the potentially informative samples are considered to be common escape genes (red) in each population, whereas the others are rare escape genes (blue) in the populations. The genes solely replicated via more than one SNPs per gene are not included. Their inclusion makes no difference to qualitative trends. The x axis refers to the count of individuals with evidence for escape in the corresponding genes. Note Xist is within the Xic domain.
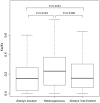
Fig. 2.
_K_a/_K_s ratios of genes in the three X-inactivation classes in the merged data set. P values indicate significance on pairwise Mann–Whitney U tests. There are 35 that always escape, 206 always inactivated, and 205 heterogeneous (N = 446). Evolutionary rates are from the human–macaque orthologous genes with numbers taken from Ensembl or from Park et al. (2010). Outliners are not shown. Transverse lines indicate the median value.
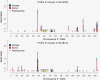
Fig. 3.
Location of escape genes and haploid expressed genes on the X chromosome of one individual of CEU (NA12004) and YRI (NA18511). Genes marked as a “potential site” are those where there is exonic heterozygozity at the DNA level and transcripts that pass the coverage threshold but that do not show evidence of escape (i.e., no evidence of biallelic expression). Those marked in blue/red show evidence of escape. The sum height of the colored bar indicates the net read depth summing over both alleles. The proportion of blue to red indicates the proportion of expression from the inactive X chromosome (blue) and the active X chromosome (we always presume the minority allele is from the inactive X chromosome). The data for the pattern of escape from the remaining individuals are shown in
supplementary figure S5
,
Supplementary Material
online.
Comment in
- X doesn't always mark the spot.
Caspermeyer J. Caspermeyer J. Mol Biol Evol. 2013 Dec;30(12):2737-8. doi: 10.1093/molbev/mst183. Epub 2013 Oct 29. Mol Biol Evol. 2013. PMID: 24174470 No abstract available.
Similar articles
- Human genes escaping X-inactivation revealed by single cell expression data.
Wainer Katsir K, Linial M. Wainer Katsir K, et al. BMC Genomics. 2019 Mar 12;20(1):201. doi: 10.1186/s12864-019-5507-6. BMC Genomics. 2019. PMID: 30871455 Free PMC article. - Strong purifying selection at genes escaping X chromosome inactivation.
Park C, Carrel L, Makova KD. Park C, et al. Mol Biol Evol. 2010 Nov;27(11):2446-50. doi: 10.1093/molbev/msq143. Epub 2010 Jun 9. Mol Biol Evol. 2010. PMID: 20534706 Free PMC article. - Global survey of escape from X inactivation by RNA-sequencing in mouse.
Yang F, Babak T, Shendure J, Disteche CM. Yang F, et al. Genome Res. 2010 May;20(5):614-22. doi: 10.1101/gr.103200.109. Epub 2010 Apr 2. Genome Res. 2010. PMID: 20363980 Free PMC article. - Genes that escape from X inactivation.
Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Berletch JB, et al. Hum Genet. 2011 Aug;130(2):237-45. doi: 10.1007/s00439-011-1011-z. Epub 2011 May 26. Hum Genet. 2011. PMID: 21614513 Free PMC article. Review. - How do genes that escape from X-chromosome inactivation contribute to Turner syndrome?
Peeters SB, Korecki AJ, Baldry SEL, Yang C, Tosefsky K, Balaton BP, Simpson EM, Brown CJ. Peeters SB, et al. Am J Med Genet C Semin Med Genet. 2019 Mar;181(1):28-35. doi: 10.1002/ajmg.c.31672. Epub 2019 Feb 19. Am J Med Genet C Semin Med Genet. 2019. PMID: 30779428 Review.
Cited by
- Analysis of sex-biased gene expression in a Eurasian admixed population.
Cheng S, Ning Z, Huang K, Yuan Y, Tan X, Pan Y, Zhang R, Tian L, Lu Y, Wang X, Lu D, Yang Y, Guan Y, Mamatyusupu D, Xu S. Cheng S, et al. Brief Bioinform. 2024 Jul 25;25(5):bbae451. doi: 10.1093/bib/bbae451. Brief Bioinform. 2024. PMID: 39293802 Free PMC article. - Sex as a Determinant of Age-Related Changes in the Brain.
Burmistrov DE, Gudkov SV, Franceschi C, Vedunova MV. Burmistrov DE, et al. Int J Mol Sci. 2024 Jun 28;25(13):7122. doi: 10.3390/ijms25137122. Int J Mol Sci. 2024. PMID: 39000227 Free PMC article. Review. - Sex-biased gene expression during neural differentiation of human embryonic stem cells.
Pottmeier P, Nikolantonaki D, Lanner F, Peuckert C, Jazin E. Pottmeier P, et al. Front Cell Dev Biol. 2024 May 1;12:1341373. doi: 10.3389/fcell.2024.1341373. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38764741 Free PMC article. - Sex differences in brain cell-type specific chromatin accessibility in schizophrenia.
Roussos P, Ma Y, Girdhar K, Hoffman G, Fullard J, Bendl J. Roussos P, et al. Res Sq [Preprint]. 2024 Apr 4:rs.3.rs-4158509. doi: 10.21203/rs.3.rs-4158509/v1. Res Sq. 2024. PMID: 38645177 Free PMC article. Preprint. - The interplay between X-chromosome functional dosage and circadian regulation in females.
Moysés-Oliveira M, Andersen ML, Tufik S. Moysés-Oliveira M, et al. Arch Womens Ment Health. 2024 Oct;27(5):845-849. doi: 10.1007/s00737-024-01452-2. Epub 2024 Apr 2. Arch Womens Ment Health. 2024. PMID: 38563984
References
- Belmont AS, Bignone F, Ts'o PO. The relative intranuclear positions of barr bodies in XXX non-transformed human fibroblasts. Exp Cell Res. 1986;165:165–179. - PubMed
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources