Lck availability during thymic selection determines the recognition specificity of the T cell repertoire - PubMed (original) (raw)
. 2013 Sep 12;154(6):1326-41.
doi: 10.1016/j.cell.2013.08.009.
Anastasia N Tikhonova, Leonid A Pobezinsky, Xuguang Tai, Motoko Y Kimura, Cécile Le Saout, Terry I Guinter, Anthony Adams, Susan O Sharrow, Günter Bernhardt, Lionel Feigenbaum, Alfred Singer
Affiliations
- PMID: 24034254
- PMCID: PMC3792650
- DOI: 10.1016/j.cell.2013.08.009
Lck availability during thymic selection determines the recognition specificity of the T cell repertoire
François Van Laethem et al. Cell. 2013.
Abstract
Thymic selection requires signaling by the protein tyrosine kinase Lck to generate T cells expressing αβ T cell antigen receptors (TCR). For reasons not understood, the thymus selects only αβTCR that are restricted by major histocompatibility complex (MHC)-encoded determinants. Here, we report that Lck proteins that were coreceptor associated promoted thymic selection of conventionally MHC-restricted TCR, but Lck proteins that were coreceptor free promoted thymic selection of MHC-independent TCR. Transgenic TCR with MHC-independent specificity for CD155 utilized coreceptor-free Lck to signal thymic selection in the absence of MHC, unlike any transgenic TCR previously described. Thus, the thymus can select either MHC-restricted or MHC-independent αβTCR depending on whether Lck is coreceptor associated or coreceptor free. We conclude that the intracellular state of Lck determines the specificity of thymic selection and that Lck association with coreceptor proteins during thymic selection is the mechanism by which MHC restriction is imposed on a randomly generated αβTCR repertoire.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
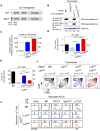
Figure 1. Effect of coreceptor-free and coreceptor-associated Lck on thymic selection
(A) hCD2-driven transgenic constructs encoding wildtype (top) and C20A/C23A mutant (bottom) Lck proteins. (B) Abrogation of Lck binding to coreceptor proteins. Thymocyte lysates were precipitated with anti-CD4 or anti-CD8α and blotted for Lck. Numbers indicate total Lck band intensities relative to B6 (set equal to 1.0). Data represent 4 experiments. (C) CD5 levels on thymocytes relative to control B6 mice (mean ± SE, n=7 mice/group). (D and E) Numbers of γδ (D) and αβ (E) LN T cells (mean ± SE, n=9 mice/group). (F) Numbers above profiles indicate thymus cellularity (mean ± SE, n=10mice/group). (G) Ligand specificity of αβT cells in mixed lymphocyte reactions as assessed byproliferation-induced CFSE dye dilution on day 4. Numbers represent percentage of cellsthat underwent at least one division. Data represent 3 experiments. **** P<0.0001; *** P<0.001; ** P<0.01. See also Figures S1 and S2.
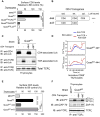
Figure 2. Lck sequestration and TCR signaling in the thymus
(A) CD5 on thymocytes relative to B6 (set at 100%). Mean ± SE, n=3 mice/group. (B) hCD2-driven transgenic constructs encoding wildtype (444) and tailless (44T) CD4 proteins named for their extra-cellular, transmembrane, and cytosolic domains. (C) Lck is sequestered away from TCR on thymocytes by the cytosolic tail of coreceptor proteins. Thymocyte lysates were precipitated by anti-CD4 or anti-TCRβ and blotted for Lck or TCRζ. Data represent 3 experiments. (D) In vitro TCR induced calcium mobilization. Thymocytes were loaded at 31oC with Indo-1 and coated with biotinylated anti-TCR (5μg/ml) either alone or together with biotinylated anti-CD4 (1μg/ml). Crosslinking by avidin (black arrow) and addition of ionomycin (red arrow) are indicated. Data represent 2 experiments. (E) In vivo signaling of thymocytes. CD5 on thymocytes is shown relative tocontrol B6 mice (mean ± SE, n=10 mice/group). (F) In vivo signaling of thymocytes assessed biochemically. Lysates wereprecipitated with anti-TCRζ and blotted for phospho-zeta, ZAP70, or TCRζ. Datarepresent 2 experiments. **** P<0.0001; *** P<0.001.
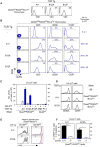
Figure 3. Thymic selection of MHC-independent αβTCR in the absence of both MHC and coreceptors
(A) TCR expression on thymocytes from transgenic host mice (QuadKORAG2KOBcl-2Tg). Black lines indicate TCR expression, grey lines indicate control staining. Numbers in panels indicate mean fluorescence intensity (MFI). Data represent 5 experiments. (B) Thymocyte profiles from host transgenic mice expressing transgenic TCR. Numbers in CD5 histograms indicate CD5 MFI. Numbers in CD69 and CCR7 histogramsindicate frequency of positive cells. Thymus cellularity is shown as mean ± SE (n=5 mice/group). (C) Numbers of αβ LN T cells in TCR transgenic mice (mean ± SE, n=5 mice/group). (D) Expression of CD62L and CD44 on αβT cells from TCR transgenic mice. Data represent 3 experiments. (E and F) Lymphopenia-induced proliferation triggered by A11 and B12A TCR is dependent on CD155 recognition. A11 and B12A αβT cells were CFSE labeled and injected (5×106 cells/mouse) into CD155+/+ or CD155−/− lymphopenic hosts for 1 week. Numbers in histograms (E) represent the percentage of cells that underwent at least two divisions and summarized in (F). Mean ± SE of 3 mice/group. **** p<0.0001; * p<0.05. See also Figure S3.
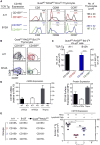
Figure 4. Identification of CD155 as the thymic selecting ligand
(A) In vivo positive selection signaling by A11 and B12A TCR requires CD155. Numbers in CD5 histograms indicate MFI. Numbers in CD69 and CCR7 histograms indicate frequency of positive cells. Thymus cellularity is shown as mean ± SE (n=5 mice/group). (B) CD69 versus CCR7 profiles of A11 and B12A thymocytes. Numbers indicate frequency of mature CD69−CCR7hi thymocytes (n=5 mice/group). (C) Numbers of αβ LN T cells (mean ± SE of 5 mice/group). (D) CD155 expression by electronically sorted thymic elements by quantitativePCR (left panel) and surface protein expression (right panel). Mean ± SE (n=3). (E) Thymic elements that induce CD155-specific thymic selection. Four groups ofmixed donor bone marrow chimeras were constructed with 1:1 mixtures of A11 andB12F donor cells and injected into irradiated host (QuadKORAG2KO) mice that expressedor lacked CD155. Numbers of A11 αβT cells in LN were determined after 8-10wks.Each circle represents one mouse (n=4). * p <0.05; **** p<0.0001; NS, not significant. See also Figure S4.
Figure 5. Positive selection signaling by MHC-independent TCR in coreceptor-sufficient mice lacking MHC
(A) Thymocyte profiles. Numbers in CD4 vs CD8α plots indicate cell frequencies. Numbers in CD5 histograms indicate MFI. Numbers in CD69 and CCR7 histograms indicate frequency of positive cells. Thymus cellularity is shown as mean ± SE (n=6 mice/group). (B) Numbers of αβ LN T cells (mean ± SE, n=6 mice/group). (C) Positive selection signaling occurs in DN thymocytes and is ligand specific. CD4 vs CD8α profiles (top) and CD5 and CCR7 expression on CD24hiDN thymocytes (bottom). Data represent 4 experiments. (D) Early positive selection signaling is TCR specific. CD5 and CCR7 expression on gated DN and DP thymocytes. Data represent 4 experiments. (E) Early positive selection signaling is inhibited by early CD4 expression. CCR7 and CD24 expression on thymocytes from B12A and B12A.444 transgenic mice. Data represent 4 individual mice/group. See also Figure S5.
Figure 6. Effect on thymic selection of coreceptor-associated and coreceptor-free Lck
(A) Numbers of αβ LN T cells in coreceptor-deficient (left side) or coreceptor-sufficient (right side) mice (mean ± SE, n=3 mice/group). (B) Numbers of αβ LN T cells in kinase-deficient mice (mean ± SE, n=3 mice/group). ND, not done. (C) Profiles of thymocytes with endogenous Lck (black lines) or Lckmut (colored lines) proteins. Numbers in CD4 vs CD8α plots indicate cell frequencies. Numbers in CD69 histograms indicate frequency of positive cells and is summarized in right panels (mean ± SE, n=3 mice/group). (D) Numbers of αβ LN T cells in TCR transgenic mice with endogenous Lck (black bar) or Lckmut (gray bar) proteins. Reconstitution by Lckmut proteins relative to endogenous Lck proteins (set at 100%) is displayed in the right panel (mean ± SE, n=3 mice/group). * p<0.05; **, p<.01; ***, p<0.001; ****, p<0.0001; NS, not significant.
Figure 7. Role of evolutionarily conserved CDR2β residues in thymic selection by an MHC-independent TCR
(A) Thymocyte profiles from MHC-deficient mice expressing A11 TCR whose TCRβ chain was wildtype or contained a point mutation in CDR2, either Y48A or E54A. Numbers in TCRβ and CD5 histograms indicate MFI. Numbers in CD69 and CCR7 histograms indicate frequency of positive cells. Numbers in CD69 vs CCR7 plots indicate frequency of mature cells. Thymus cellularity is shown as mean ± SE (n=4 mice/group). (B) Assessment of in vivo signaling of thymocytes. Unstimulated thymocyte lysates were precipitated with anti-TCRζ and blotted for phospho-zeta, ZAP70, or TCRζ. Data represent 2 experiments. (C) LN αβT cell numbers (mean ± SE, n=4 mice/group). * p<0.05; ****, p<0.0001.
Similar articles
- Subcellular distribution of Lck during CD4 T-cell maturation in the thymic medulla regulates the T-cell activation threshold.
Stephen TL, Wilson BS, Laufer TM. Stephen TL, et al. Proc Natl Acad Sci U S A. 2012 May 8;109(19):7415-20. doi: 10.1073/pnas.1119272109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529380 Free PMC article. - Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC.
Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, Adoro S, Adams A, Sharrow SO, Feigenbaum L, Singer A. Van Laethem F, et al. Immunity. 2007 Nov;27(5):735-50. doi: 10.1016/j.immuni.2007.10.007. Immunity. 2007. PMID: 18023370 - Autonomous maturation of alpha/beta T lineage cells in the absence of COOH-terminal Src kinase (Csk).
Schmedt C, Tarakhovsky A. Schmedt C, et al. J Exp Med. 2001 Apr 2;193(7):815-26. doi: 10.1084/jem.193.7.815. J Exp Med. 2001. PMID: 11283154 Free PMC article. - CD4 and CD8: hogging all the Lck.
Kappes DJ. Kappes DJ. Immunity. 2007 Nov;27(5):691-3. doi: 10.1016/j.immuni.2007.11.002. Immunity. 2007. PMID: 18031691 Review. - Thymic Origins of T Cell Receptor Alloreactivity.
Brzostek J, Gascoigne NRJ. Brzostek J, et al. Transplantation. 2017 Jul;101(7):1535-1541. doi: 10.1097/TP.0000000000001654. Transplantation. 2017. PMID: 28114168 Review.
Cited by
- The partitioning of TCR repertoires by thymic selection.
Lo WL, Huseby ES. Lo WL, et al. J Exp Med. 2024 Oct 7;221(10):e20230897. doi: 10.1084/jem.20230897. Epub 2024 Aug 21. J Exp Med. 2024. PMID: 39167074 Free PMC article. Review. - A Story of Kinases and Adaptors: The Role of Lck, ZAP-70 and LAT in Switch Panel Governing T-Cell Development and Activation.
Fernández-Aguilar LM, Vico-Barranco I, Arbulo-Echevarria MM, Aguado E. Fernández-Aguilar LM, et al. Biology (Basel). 2023 Aug 24;12(9):1163. doi: 10.3390/biology12091163. Biology (Basel). 2023. PMID: 37759563 Free PMC article. Review. - The H2-A Class II molecule α/β-chain _cis-_mismatch severely affects cell surface expression, selection of conventional CD4+ T cells and protection against TB infection.
Logunova N, Kapina M, Kondratieva E, Apt A. Logunova N, et al. Front Immunol. 2023 Jun 22;14:1183614. doi: 10.3389/fimmu.2023.1183614. eCollection 2023. Front Immunol. 2023. PMID: 37426653 Free PMC article. - The kinase occupancy of T cell coreceptors reconsidered.
Mørch AM, Schneider F, Jenkins E, Santos AM, Fraser SE, Davis SJ, Dustin ML. Mørch AM, et al. Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2213538119. doi: 10.1073/pnas.2213538119. Epub 2022 Dec 1. Proc Natl Acad Sci U S A. 2022. PMID: 36454761 Free PMC article. - Integrative and Comprehensive Pan-Cancer Analysis of Lymphocyte-Specific Protein Tyrosine Kinase in Human Tumors.
Han M, Li Y, Guo Y, Zhu W, Jiang J. Han M, et al. Int J Mol Sci. 2022 Nov 13;23(22):13998. doi: 10.3390/ijms232213998. Int J Mol Sci. 2022. PMID: 36430477 Free PMC article.
References
- Abraham N, Miceli MC, Parnes JR, Veillette A. Enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Nature. 1991;350:62–66. - PubMed
- Acuto O, Cantrell D. T cell activation and the cytoskeleton. Annu Rev Immunol. 2000;18:165–184. - PubMed
- Chan AC, Iwashima M, Turck CW, Weiss A. ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell. 1992;71:649–662. - PubMed
- Chien YH, Konigshofer Y. Antigen recognition by gammadelta T cells. Immunological reviews. 2007;215:46–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous