Protein similarity networks reveal relationships among sequence, structure, and function within the Cupin superfamily - PubMed (original) (raw)
Protein similarity networks reveal relationships among sequence, structure, and function within the Cupin superfamily
Richard Uberto et al. PLoS One. 2013.
Abstract
The cupin superfamily is extremely diverse and includes catalytically inactive seed storage proteins, sugar-binding metal-independent epimerases, and metal-dependent enzymes possessing dioxygenase, decarboxylase, and other activities. Although numerous proteins of this superfamily have been structurally characterized, the functions of many of them have not been experimentally determined. We report the first use of protein similarity networks (PSNs) to visualize trends of sequence and structure in order to make functional inferences in this remarkably diverse superfamily. PSNs provide a way to visualize relatedness of structure and sequence among a given set of proteins. Structure- and sequence-based clustering of cupin members reflects functional clustering. Networks based only on cupin domains and networks based on the whole proteins provide complementary information. Domain-clustering supports phylogenetic conclusions that the N- and C-terminal domains of bicupin proteins evolved independently. Interestingly, although many functionally similar enzymatic cupin members bind the same active site metal ion, the structure and sequence clustering does not correlate with the identity of the bound metal. It is anticipated that the application of PSNs to this superfamily will inform experimental work and influence the functional annotation of databases.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
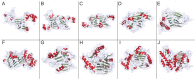
Figure 1. Structures of representative of members of the cupin superfamily.
A. oxalate oxidase (PDB code: 2et1) , B. oxalate decarboxylase (PDB code: 1uw8) , C. seed storage protein Ara h (PDB code: 3s7i) , D. NovW, a 4-keto-6-deoxy sugar epimerase (PDB code: 2c0z) , E. cysteine dioxygenase (PDB code: 2q4s) , F. phosphomannose isomerase (PDB code: 1 pmi) G. acireductone dioxygenase (PDB code: 1 zrr) H. taurine/alpha-ketoglutarate dioxygenase (PDB code: 1os7) , I. hypoxia-inducible factor 1-alpha inhibitor (PBD code: 2y0i) , J. lysine-specific demethylase 6B (PDB code: 2 xue) . β-sheets are shown in green, α-helices are shown in red, and random coils are shown in grey. Spheres represent bound metal ions. Figures were generated using Pymol (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Figure 2. Metal-binding sites of representative members of the cupin superfamily.
A. Mn ion of oxalate oxidase coordinated by His88, His90, Glu95, and His137 (PDB code: 2et1) , B. N-terminal Mn ion of oxalate decarboxylase coordinated by His95, His97, Glu101, and His140 (PDB code: 1uw8) , C. C-terminal Mn ion of oxalate decarboxylase coordinated by His273, His275, Glu280, and His319 (PDB code: 1uw8) , D. Ni ion of cysteine dioxygenase coordinated by His86, His88, and His140 (PDB code: 2q4s) , E. Zn ion of phosphomannose isommerase coordinated by Gln111, His113, Glu138, and His285 (PDB code: 1 pmi) F. Ni ion of acireductone dioxygenase coordinated by His96, His98, Glu102, and His140 (PDB code: 1 zrr) , G. Fe ion of taurine/alphaketoglutarate dioxygenase coordinated by His99, Asp101, and His255 (PDB code: 1os7) , H. Fe ion of hypoxia-inducible factor 1-alpha inhibitor coordinated by His199, Asp201, and His279 (PBD code: 2y0i) , I. Fe ion of lysine-specific demethylase 6B coordinated by His1390, Glu1392, and His1470 (PDB code: 2 xue) , J. Zn ion of lysine-specific demethylase 6B coordinated by Cys1575, Cys1578, Cys1602, and Cys1605 (not part of the cupin domain) (PDB code: 2 xue) . β-sheets are shown in green, α-helices are shown in red, and random coils are shown in grey. Spheres represent bound metal ions. Figures were generated using Pymol (The PyMOL Molecular Graphics System, Schrödinger, LLC).
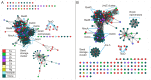
Figure 3. Structure similarity networks of the cupin protein stuctures colored by metal ligand.
Pairwise similarities for a non-redundant set of 183 structures from the Pfam cupin clan (CL0029) were calculated using TM-align . Each node represents a structure. Nodes were arranged using the yfiles organic layout of of Cytoscape version 3.0. A. Edges between nodes were drawn only if the average TM-score >0.53 for that edge. At this cutoff, the average r.m.s.d. is 2.91 Å with an average of 158.0 Cα atoms aligned. B. Edges between nodes were drawn only if the average TM-score >0.65 for that edge. At this cutoff, the average r.m.s.d. is 2.44 Å with an average of 185.4 Cα atoms aligned.
Figure 4. Structure similarity networks of cupin domains colored by metal ligand.
Pairwise similarities for a non-redundant set of 213 domains from the Pfam cupin clan (CL0029) were calculated using TM-align . Each node represents a domain. Nodes were arranged using the yfiles organic layout of of Cytoscape version 3.0. A. Edges between nodes were drawn only if the average TM-score >0.53 for that edge. At this cutoff, the average r.m.s.d. is 1.73 Å with 74.2 Cα atoms aligned. B. Edges between nodes were drawn only if the average TM-score >0.65 for that edge. At this cutoff, the average r.m.s.d. is 1.42 Å with 80.1 Cαatoms aligned.
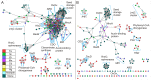
Figure 5. Structure similarity networks colored by species and function.
This is the same network as in 3B but colored according to A. species and B. function.
Figure 6. Sequence similarity networks colored by metal ligand.
Networks were generated by all-by all BLAST comparisons of the 183 sequences corresponding to the unique cupin structures shown in Figure 3. Nodes were arranged using the yfiles organic layout of of Cytoscape version 3.0. A. Edges between nodes are drawn only if the E-value is better than of 1E-3.5. At this cutoff, edges at this threshold represent alignments with a median 32.1% identity over 93 residues. B. Edges between nodes are drawn only if the E-value is better than of 1E-6.0. At this cutoff, edges at this threshold represent alignments with a median 36.2% identity over 185 residues.
Similar articles
- [Evolution of seed storage globulins and cupin superfamily].
Shutov AD, Kakhovskaia IA. Shutov AD, et al. Mol Biol (Mosk). 2011 Jul-Aug;45(4):579-85. Mol Biol (Mosk). 2011. PMID: 21954589 Review. Russian. - D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ribulose-phosphate binding (beta/alpha)8-barrel superfamily.
Akana J, Fedorov AA, Fedorov E, Novak WR, Babbitt PC, Almo SC, Gerlt JA. Akana J, et al. Biochemistry. 2006 Feb 28;45(8):2493-503. doi: 10.1021/bi052474m. Biochemistry. 2006. PMID: 16489742 - Sequence analysis of the cupin gene family in Synechocystis PCC6803.
Dunwell JM. Dunwell JM. Microb Comp Genomics. 1998;3(2):141-8. doi: 10.1089/omi.1.1998.3.141. Microb Comp Genomics. 1998. PMID: 9697098 - Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold.
Agarwal G, Rajavel M, Gopal B, Srinivasan N. Agarwal G, et al. PLoS One. 2009 May 29;4(5):e5736. doi: 10.1371/journal.pone.0005736. PLoS One. 2009. PMID: 19478949 Free PMC article. - Thiol dioxygenases: unique families of cupin proteins.
Stipanuk MH, Simmons CR, Karplus PA, Dominy JE Jr. Stipanuk MH, et al. Amino Acids. 2011 Jun;41(1):91-102. doi: 10.1007/s00726-010-0518-2. Epub 2010 Mar 1. Amino Acids. 2011. PMID: 20195658 Free PMC article. Review.
Cited by
- Overproduction, crystallization and X-ray diffraction data analysis of ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis.
Kobus S, Widderich N, Hoeppner A, Bremer E, Smits SH. Kobus S, et al. Acta Crystallogr F Struct Biol Commun. 2015 Aug;71(Pt 8):1027-32. doi: 10.1107/S2053230X15011115. Epub 2015 Jul 28. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26249694 Free PMC article. - Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Widderich N, Kobus S, Höppner A, Riclea R, Seubert A, Dickschat JS, Heider J, Smits SH, Bremer E. Widderich N, et al. PLoS One. 2016 Mar 17;11(3):e0151285. doi: 10.1371/journal.pone.0151285. eCollection 2016. PLoS One. 2016. PMID: 26986827 Free PMC article. - Discovery of a Chemical Probe Bisamide (CCT251236): An Orally Bioavailable Efficacious Pirin Ligand from a Heat Shock Transcription Factor 1 (HSF1) Phenotypic Screen.
Cheeseman MD, Chessum NE, Rye CS, Pasqua AE, Tucker MJ, Wilding B, Evans LE, Lepri S, Richards M, Sharp SY, Ali S, Rowlands M, O'Fee L, Miah A, Hayes A, Henley AT, Powers M, Te Poele R, De Billy E, Pellegrino L, Raynaud F, Burke R, van Montfort RL, Eccles SA, Workman P, Jones K. Cheeseman MD, et al. J Med Chem. 2017 Jan 12;60(1):180-201. doi: 10.1021/acs.jmedchem.6b01055. Epub 2016 Dec 22. J Med Chem. 2017. PMID: 28004573 Free PMC article. - The origin and evolution of ribonucleotide reduction.
Lundin D, Berggren G, Logan DT, Sjöberg BM. Lundin D, et al. Life (Basel). 2015 Feb 27;5(1):604-36. doi: 10.3390/life5010604. Life (Basel). 2015. PMID: 25734234 Free PMC article. Review. - Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency.
Chen M, Wang C, Bao H, Chen H, Wang Y. Chen M, et al. Mol Genet Genomics. 2016 Aug;291(4):1663-80. doi: 10.1007/s00438-016-1210-3. Epub 2016 May 2. Mol Genet Genomics. 2016. PMID: 27138920
References
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65: 7–17. - PubMed
- Dunwell JM, Gane PJ (1998) Microbial relatives of seed storage proteins: conservation of motifs in a functionally diverse superfamily of enzymes. J Mol Evol 46: 147–154. - PubMed
- Dunwell JM (1998) Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol Genet Eng Rev 15: 1–32. - PubMed
- Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW (2001) Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci 26: 740–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by the National Science Foundation (MCB-1041912) to EWM (http://www.nsf.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources