Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory - PubMed (original) (raw)
Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory
Elias M Puchner et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Cells tightly regulate trafficking of intracellular organelles, but a deeper understanding of this process is technically limited by our inability to track the molecular composition of individual organelles below the diffraction limit in size. Here we develop a technique for intracellularly calibrated superresolution microscopy that can measure the size of individual organelles as well as accurately count absolute numbers of molecules, by correcting for undercounting owing to immature fluorescent proteins and overcounting owing to fluorophore blinking. Using this technique, we characterized the size of individual vesicles in the yeast endocytic pathway and the number of accessible phosphatidylinositol 3-phosphate binding sites they contain. This analysis reveals a characteristic vesicle maturation trajectory of composition and size with both stochastic and regulated components. The trajectory displays some cell-to-cell variability, with smaller variation between organelles within the same cell. This approach also reveals mechanistic information on the order of events in this trajectory: Colocalization analysis with known markers of different vesicle maturation stages shows that phosphatidylinositol 3-phosphate production precedes fusion into larger endosomes. This single-organelle analysis can potentially be applied to a range of small organelles to shed light on their precise composition/structure relationships, the dynamics of their regulation, and the noise in these processes.
Keywords: GTPases; PALM; endocytosis; phosphoinositides; single-molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
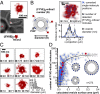
Intracellular calibration of superresolution microscopy allows counting of biomolecules in subdiffraction-limit structures. (A) Intracellular structure size and number of biomolecules span several orders of magnitude. (B) Resolution and counting accuracy are determined using constructs with different mEos2 stoichiometry. (Left) Superresolution images of calibration constructs (red) are superimposed on transmitted light images of yeast cells (gray). Single (1×, Top), double (2×, Middle) and triple (3×, Bottom) repeats of mEos2 were constitutively expressed as fusions with the membrane-localized PH domain of Plcδ. (Center) The magnified fields depict uncorrected (+) and corrected (X) single molecule positions. Each molecule and localization is color-coded by frame number. The number of molecules per cluster is fitted to a binomial distribution B(N,F) (red line). (Right) Pair-correlation functions of corrected images reflect the average distance between molecules, which is constant for the single mEos2 repeat and peaked for the 2× and 3× repeat owing to colocalization. The peak width of corrected 2× and 3× data are narrower than for uncorrected data (only shown for 1×) and reflects the increase in resolution by combining photons from all fluorescent bursts to the molecule's position. (C) Summary of calibration results. The ensemble values of corrected molecules per cell exhibit a linear relation with an expected two- and threefold higher number of mEos2 molecules of the 2× and 3× repeat compared with the single repeat. The table summarizes fraction of observable mEos2 molecules F from the single molecule counting data.
Fig. 2.
Vesicles in the endocytic pathway show characteristic relationship between vesicle size and PI3P content. (A) A tandem repeat of the FYVE domain of EEA1 fused to mEos2 serves as a specific probe to detect accessible PI3P binding sites on endocytic/endosomal vesicles. mEos2 (red) localizes to distinct, circular vesicles within a yeast cell (gray). (B) By applying our calibrated superresolution microscopy approach, the number of mEos2 molecules (N) bound to PI3P on each vesicle is determined (x’s). The molecular distribution exhibits a circular shape (two peaks in a 1D projection). The diameter of vesicles (D) is determined by measuring the spatial spread of molecules. (C) Vesicles display variability in PI3P content and size. Representative vesicles are shown displaying the expected circular shape. The histogram of vesicle diameters exhibits a fitted maximum at 82 nm. (D) PI3P content and vesicle surface area fall on a characteristic curve. Vesicles with fewer than 100 molecules display relatively tight size regulation, whereas structures above this threshold are significantly larger. Vesicles from each part of this characteristic distribution can be found within one individual cell (red). This path is also displayed as an exponential function (black line) fitted to box-smoothed data with 95% confidence envelope.
Fig. 3.
Counting of PI3P-binding sites is validated with conventional fluorescence. (A) Schematic for validation. One PI3P probe (A) is tagged with mEos2, allowing for high-resolution measurements of structure and quantification. The other (B) is fused to GFP, which can be quantified by calculating the total fluorescence intensity. (B) Quantified conventional fluorescence validates superresolution counting. FYVE–mEos2 and FYVE–GFP constructs were expressed in the same cells. A colocalization image shows transmitted light image (gray), mEos2 (red), and GFP (green). The number of mEos2 molecules determined in the red superresolution channel and the amount of GFP quantified by total fluorescence intensity on each vesicle is tightly correlated (Pearson’s coefficient, ρ= 0.91).
Fig. 4.
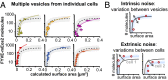
Variation in the relationship between PI3P-binding site number and vesicle size is due to both intrinsic and extrinsic noise in the endocytic pathway. (A) Measurements made on several vesicles within one individual cell display variation around the characteristic curve. Vesicles from all regions of the PI3P number-size distribution can be found within the same cell. Vesicles within the same cell may be located above (red) or below (pink) the characteristic curve (dotted line, 95% confidence interval as gray shading). (B) Schematics showing two components of noise in PI3P content and size. (Upper) Intrinsic noise is due to a variation between vesicles within a cell and (Lower) extrinsic noise is due to systematic variations between cells.
Fig. 5.
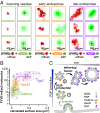
Benchmarking with endocytic/endosomal landmark proteins reveals distinct phases of PI3P production and vesicle fusion. (A) Colocalization of vesicles with landmark proteins of the endocytic pathway. Representative superresolution images (red, left column of each pair) and GFP images (green, right column of each pair) for each class of protein are shown. (B) Colocalization with endocytic pathway proteins reveals distinct subclasses of vesicles. FYVE–mEos2 colocalizes with GFP-tagged clathrin (green) and GTPases Vps21p (orange) and Ypt7p (purple). For each dataset, the median, upper, and lower quartiles are displayed. (C) Model of PI3P production on vesicles in endocytic pathway followed by fusion. Phases of endocytic/endosomal protein association, enzymatic production of PI3P, and fusion to early and late endosomes (1–3) are described in B.
Similar articles
- Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry.
Cuesta-Geijo MA, Galindo I, Hernáez B, Quetglas JI, Dalmau-Mena I, Alonso C. Cuesta-Geijo MA, et al. PLoS One. 2012;7(11):e48853. doi: 10.1371/journal.pone.0048853. Epub 2012 Nov 1. PLoS One. 2012. PMID: 23133661 Free PMC article. - Phosphorylation of the Rab exchange factor Sec2p directs a switch in regulatory binding partners.
Stalder D, Mizuno-Yamasaki E, Ghassemian M, Novick PJ. Stalder D, et al. Proc Natl Acad Sci U S A. 2013 Dec 10;110(50):19995-20002. doi: 10.1073/pnas.1320029110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248333 Free PMC article. - The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles.
Poupon V, Stewart A, Gray SR, Piper RC, Luzio JP. Poupon V, et al. Mol Biol Cell. 2003 Oct;14(10):4015-27. doi: 10.1091/mbc.e03-01-0040. Epub 2003 Jul 11. Mol Biol Cell. 2003. PMID: 14517315 Free PMC article. - Who's in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond.
Borchers AC, Langemeyer L, Ungermann C. Borchers AC, et al. J Cell Biol. 2021 Sep 6;220(9):e202105120. doi: 10.1083/jcb.202105120. Epub 2021 Aug 12. J Cell Biol. 2021. PMID: 34383013 Free PMC article. Review. - Chasing Uptake: Super-Resolution Microscopy in Endocytosis and Phagocytosis.
Baranov MV, Olea RA, van den Bogaart G. Baranov MV, et al. Trends Cell Biol. 2019 Sep;29(9):727-739. doi: 10.1016/j.tcb.2019.05.006. Epub 2019 Jun 18. Trends Cell Biol. 2019. PMID: 31227311 Review.
Cited by
- Quantifying Protein Copy Number in Super Resolution Using an Imaging-Invariant Calibration.
Cella Zanacchi F, Manzo C, Magrassi R, Derr ND, Lakadamyali M. Cella Zanacchi F, et al. Biophys J. 2019 Jun 4;116(11):2195-2203. doi: 10.1016/j.bpj.2019.04.026. Epub 2019 May 3. Biophys J. 2019. PMID: 31103226 Free PMC article. - Endosomal Interactions during Root Hair Growth.
von Wangenheim D, Rosero A, Komis G, Šamajová O, Ovečka M, Voigt B, Šamaj J. von Wangenheim D, et al. Front Plant Sci. 2016 Jan 29;6:1262. doi: 10.3389/fpls.2015.01262. eCollection 2015. Front Plant Sci. 2016. PMID: 26858728 Free PMC article. - In cellulo evaluation of phototransformation quantum yields in fluorescent proteins used as markers for single-molecule localization microscopy.
Avilov S, Berardozzi R, Gunewardene MS, Adam V, Hess ST, Bourgeois D. Avilov S, et al. PLoS One. 2014 Jun 10;9(6):e98362. doi: 10.1371/journal.pone.0098362. eCollection 2014. PLoS One. 2014. PMID: 24915511 Free PMC article. - Unscrambling fluorophore blinking for comprehensive cluster detection via photoactivated localization microscopy.
Platzer R, Rossboth BK, Schneider MC, Sevcsik E, Baumgart F, Stockinger H, Schütz GJ, Huppa JB, Brameshuber M. Platzer R, et al. Nat Commun. 2020 Oct 5;11(1):4993. doi: 10.1038/s41467-020-18726-9. Nat Commun. 2020. PMID: 33020470 Free PMC article. - Challenges facing quantitative large-scale optical super-resolution, and some simple solutions.
Dankovich TM, Rizzoli SO. Dankovich TM, et al. iScience. 2021 Feb 3;24(3):102134. doi: 10.1016/j.isci.2021.102134. eCollection 2021 Mar 19. iScience. 2021. PMID: 33665555 Free PMC article. Review.
References
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. - PubMed
- Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. - PubMed
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440(7082):358–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM55040/GM/NIGMS NIH HHS/United States
- 5F32GM093475/GM/NIGMS NIH HHS/United States
- P50GM081879/GM/NIGMS NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- R01 GM062583/GM/NIGMS NIH HHS/United States
- DP2 OD008479/OD/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM62583/GM/NIGMS NIH HHS/United States
- PN2 EY016546/EY/NEI NIH HHS/United States
- 1DP2OD008479/OD/NIH HHS/United States
- R01 GM055040/GM/NIGMS NIH HHS/United States
- F32 GM093475/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases