Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop - PubMed (original) (raw)
Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop
Phanourios Tamamis et al. Biophys J. 2013.
Abstract
HIV-1 cell entry is initiated by the interaction of the viral envelope glycoprotein gp120 with CD4, and chemokine coreceptors CXCR4 and CCR5. The molecular recognition of CXCR4 or CCR5 by the HIV-1 gp120 is mediated through the V3 loop, a fragment of gp120. The binding of the V3 loop to CXCR4 or CCR5 determines the cell tropism of HIV-1 and constitutes a key step before HIV-1 cell entry. Thus, elucidating the molecular recognition of CXCR4 by the V3 loop is important for understanding HIV-1 viral infectivity and tropism, and for the design of HIV-1 inhibitors. We employed a comprehensive set of computational tools, predominantly based on free energy calculations and molecular-dynamics simulations, to investigate the molecular recognition of CXCR4 by a dual tropic V3 loop. We report what is, to our knowledge, the first HIV-1 gp120 V3 loop:CXCR4 complex structure. The computationally derived structure reveals an abundance of polar and nonpolar intermolecular interactions contributing to the HIV-1 gp120:CXCR4 binding. Our results are in remarkable agreement with previous experimental findings. Therefore, this work sheds light on the functional role of HIV-1 gp120 V3 loop and CXCR4 residues associated with HIV-1 coreceptor activity.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
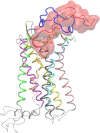
Figure 1
Molecular graphics image of the entire simulation system corresponding to the complex with the lowest average binding free energy. The V3 loop is shown in tube and transparent surface representation in red color, and its 16–20 residue moiety is shown in thick tube representation. The CXCR4 is shown in cartoon representation, and the coloring used for different protein domains is as follows: (blue) N-terminal domain; (green) intramembrane helix 1 (IH1); (light gray) intracellular loop 1 (ICL1); (purple) IH2L; (light gray) ECL1; (yellow) IH3; (light gray) ICL 2; (medium gray) IH4; (ochre) ECL2; (pink) IH5; (light gray) ICL3; (cyan) IH6; (lime) ECL3; (orange) IH7; and (light gray) C-terminal domain. (van der Waals sphere) N-terminal C_α_ atom of CXCR4. (Thick transparent licorice representation) V3 loop disulfide bridge. The definition of CXCR4 and V3 loop domains is presented in the Supporting Material (colors appear in the online version only).
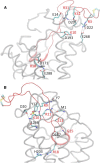
Figure 2
Molecular graphics images of important polar interactions corresponding to the complex with the lowest average binding free energy. Panels (A) and (B) depict the salt bridges and the most important hydrogen bonds, respectively, according to residue pair-wise interaction free energies. Panel (B) is rotated by approximately 180° with respect to (A) around the z (membrane) axis. The V3 loop is shown in tube and in red color, and its 16–20 residue moiety is shown in fat tube representation. The CXCR4 is shown in light gray transparent tube representation. The salt bridge and hydrogen bonds in panels (A) and (B) are denoted in dashed lines and the participating V3 loop and CXCR4 residue moieties are shown in licorice; V3 loop and CXCR4 residues are annotated in red and black, color respectively. Hydrogen atoms are omitted for clarity and the V3 loop disulfide bridge is shown in fat transparent licorice representation. (colors appear in the online version only).
Similar articles
- Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop.
Tamamis P, Floudas CA. Tamamis P, et al. PLoS One. 2014 Apr 24;9(4):e95767. doi: 10.1371/journal.pone.0095767. eCollection 2014. PLoS One. 2014. PMID: 24763408 Free PMC article. - HIV-1 gp120-CXCR4 recognition probed with synthetic nanomolar affinity D-peptides containing fragments of gp120 V3 loop.
Zhu R, Meng Q, Zhang H, Zhang G, Huang LSM, Xu Y, Schooley RT, An J, Huang Z. Zhu R, et al. Eur J Med Chem. 2022 Dec 15;244:114797. doi: 10.1016/j.ejmech.2022.114797. Epub 2022 Sep 28. Eur J Med Chem. 2022. PMID: 36270088 Free PMC article. - Role of the HIV type 1 glycoprotein 120 V3 loop in determining coreceptor usage.
Verrier F, Borman AM, Brand D, Girard M. Verrier F, et al. AIDS Res Hum Retroviruses. 1999 May 20;15(8):731-43. doi: 10.1089/088922299310827. AIDS Res Hum Retroviruses. 1999. PMID: 10357469 - HIV-1 gp120 V3 loop for structure-based drug design.
Sirois S, Sing T, Chou KC. Sirois S, et al. Curr Protein Pept Sci. 2005 Oct;6(5):413-22. doi: 10.2174/138920305774329359. Curr Protein Pept Sci. 2005. PMID: 16248793 Review. - Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.
Panos G, Watson DC. Panos G, et al. Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
Cited by
- Highly Accurate and Efficient Deep Learning Paradigm for Full-Atom Protein Loop Modeling with KarmaLoop.
Wang T, Zhang X, Zhang O, Chen G, Pan P, Wang E, Wang J, Wu J, Zhou D, Wang L, Jin R, Chen S, Shen C, Kang Y, Hsieh CY, Hou T. Wang T, et al. Research (Wash D C). 2024 Jul 25;7:0408. doi: 10.34133/research.0408. eCollection 2024. Research (Wash D C). 2024. PMID: 39055686 Free PMC article. - Identifying and Assessing Putative Allosteric Sites and Modulators for CXCR4 Predicted through Network Modeling and Site Identification by Ligand Competitive Saturation.
Inan T, Flinko R, Lewis GK, MacKerell AD Jr, Kurkcuoglu O. Inan T, et al. J Phys Chem B. 2024 May 30;128(21):5157-5174. doi: 10.1021/acs.jpcb.4c00925. Epub 2024 Apr 22. J Phys Chem B. 2024. PMID: 38647430 Free PMC article. - Distinct genetic clusters in HIV-1 CRF01_AE-infected patients induced variable degrees of CD4+ T-cell loss.
Li K, Chen H, Li J, Feng Y, Liang S, Rashid A, Liu M, Li S, Chu Q, Ruan Y, Xing H, Lan G, Qiao W, Shao Y. Li K, et al. mBio. 2024 Mar 13;15(3):e0334923. doi: 10.1128/mbio.03349-23. Epub 2024 Feb 22. mBio. 2024. PMID: 38385695 Free PMC article. - CXCR4 Is a Potential Target for Anti-HIV Gene Therapy.
Prokopovich AK, Litvinova IS, Zubkova AE, Yudkin DV. Prokopovich AK, et al. Int J Mol Sci. 2024 Jan 18;25(2):1187. doi: 10.3390/ijms25021187. Int J Mol Sci. 2024. PMID: 38256260 Free PMC article. Review. - Spermine and spermidine bind CXCR4 and inhibit CXCR4- but not CCR5-tropic HIV-1 infection.
Harms M, Smith N, Han M, Groß R, von Maltitz P, Stürzel C, Ruiz-Blanco YB, Almeida-Hernández Y, Rodriguez-Alfonso A, Cathelin D, Caspar B, Tahar B, Sayettat S, Bekaddour N, Vanshylla K, Kleipass F, Wiese S, Ständker L, Klein F, Lagane B, Boonen A, Schols D, Benichou S, Sanchez-Garcia E, Herbeuval JP, Münch J. Harms M, et al. Sci Adv. 2023 Jul 7;9(27):eadf8251. doi: 10.1126/sciadv.adf8251. Epub 2023 Jul 5. Sci Adv. 2023. PMID: 37406129 Free PMC article.
References
- Chan D.C., Fass D., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. - PubMed
- Kwong P.D., Wyatt R., Hendrickson W.A. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials