Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection - PubMed (original) (raw)
. 2013 Oct 24;502(7472):559-62.
doi: 10.1038/nature12542. Epub 2013 Sep 18.
Affiliations
- PMID: 24048477
- PMCID: PMC3808269
- DOI: 10.1038/nature12542
Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection
Caroline Goujon et al. Nature. 2013.
Abstract
Animal cells harbour multiple innate effector mechanisms that inhibit virus replication. For the pathogenic retrovirus human immunodeficiency virus type 1 (HIV-1), these include widely expressed restriction factors, such as APOBEC3 proteins, TRIM5-α, BST2 (refs 4, 5) and SAMHD1 (refs 6, 7), as well as additional factors that are stimulated by type 1 interferon (IFN). Here we use both ectopic expression and gene-silencing experiments to define the human dynamin-like, IFN-induced myxovirus resistance 2 (MX2, also known as MXB) protein as a potent inhibitor of HIV-1 infection and as a key effector of IFN-α-mediated resistance to HIV-1 infection. MX2 suppresses infection by all HIV-1 strains tested, has equivalent or reduced effects on divergent simian immunodeficiency viruses, and does not inhibit other retroviruses such as murine leukaemia virus. The Capsid region of the viral Gag protein dictates susceptibility to MX2, and the block to infection occurs at a late post-entry step, with both the nuclear accumulation and chromosomal integration of nascent viral complementary DNA suppressed. Finally, human MX1 (also known as MXA), a closely related protein that has long been recognized as a broadly acting inhibitor of RNA and DNA viruses, including the orthomyxovirus influenza A virus, does not affect HIV-1, whereas MX2 is ineffective against influenza virus. MX2 is therefore a cell-autonomous, anti-HIV-1 resistance factor whose purposeful mobilization may represent a new therapeutic approach for the treatment of HIV/AIDS.
Figures
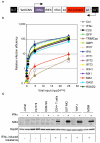
Figure 1. Human MX2 is a potent inhibitor of HIV-1 infection
A. Schematic representation of the EasiLV (
E
2-crimson
a
nti
s
ense
i
nducible
l
entiviral
v
ector) system. pEasiLV-MCS contains an internal antisense and Tet-inducible expression cassette driving expression of a tricistronic RNA encoding the cDNA of interest, the rtTA3 transcription transactivator and the E2-crimson indicator gene (see supplemental methods for details). B. Candidate cDNA screen in U87-MG/CD4/CXCR4 cells. U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing the different candidate cDNAs, CD8 (negative control), GFP (negative control) or TRIMCyp (positive control) cDNAs and treated with doxycycline for 48 h, left untransduced (Ctrl) or treated with 1000 U/ml IFNα for 24 h prior to HIV-1 infection. The cells were infected with increasing viral inputs of NL4-3/nef-IRES-renilla (0.04 to 25 ng p24Gag) and infection efficiency was monitored 48 h later by measuring renilla activity. Mean relative infection efficiencies with standard deviations from four independent experiments are shown. C. Immunoblot analysis of MX2 protein levels in control (Ctrl) and IFNα-treated Jurkat, HUT78, CEM-SS, primary CD4+ T cells, U87-MG, THP-1 and MDMs; Hsp90 served as a loading control. The IFNα-induced resistance phenotype of each cell type is shown underneath (−no resistance; + resistance).
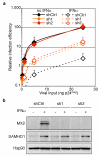
Figure 2. MX2 is required for effective IFNα-induced suppression of HIV-1
A. U87-MG/CD4/CXCR4 cells expressing a control shRNA or 2 different shRNAs targeting MX2 were treated or not with IFNα (500 U/ml) for 24 h. Cells were infected with 5 different doses of NL4-3/nef-IRES-renilla (0.04 to 25 ng p24Gag) for 48 h, and renilla activity was measured. Mean relative infection efficiencies from two independent experiments are shown. B. Immunoblot analysis of parallel samples from A. Protein levels of MX2 and SAMHD1 (positive control for IFNα induction) were determined and Hsp90 served as a loading control.
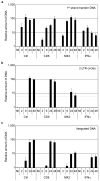
Figure 3. MX2 inhibits the nuclear accumulation and integration of HIV-1 reverse transcripts
U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8 or MX2 and treated with doxycycline for 48 h, left untransduced (Ctrl) or treated with IFNα for 24 h prior to infection. The cells were either not infected (NI) or challenged with 10 ng p24Gag HIV-1IIIB and harvested at 2, 6, 24 or 48 h post infection for DNA extraction and qPCR analysis of minus strand DNA (1st strand transfer, panel A), 2-LTR circle DNA (panel B), and integrated proviral DNA (panel C). Mean values of relative amounts of DNA (normalised to Ctrl at 48 h) from three independent experiments are shown. The detection limit for 2-LTR circle qPCR was 10 copies per reaction, which corresponds to ~6% relative copies as indicated on the graph by a dashed grey line. p24Gag expression was also determined at 48 h in parallel samples to monitor productive infection (Fig S3).
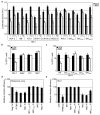
Figure 4. Viral substrates for the human MX1 and MX2 proteins
A. U87-MG/LTR-Luc cells were transduced with EasiLV expressing CD8 or MX2. Cells were infected with 2 doses (1 and 10, corresponding to 50 and 500 pg RT) of VSV G pseudotyped-HIV-1NL4-3, HIV-1IIIB, HIV-1YU-2, HIV-1CH077.t, HIV-1CH106.c, HIV-1REJO.c, HIV-2ROD10, SIVMAC239, SIVAGMTAN and SIVMND121. Luciferase activity was measured at 48 h. Mean values for three independent experiments are shown. B. CD8 or MX2 expressing U87-MG cells were challenged with HIV-1-, EIAV-, FIV-, and MLV-based retroviral vectors expressing GFP at an MOI of 0.25. The percent of GFP-expressing cells was evaluated by flow cytometry. Mean percentages of transduced cells for four independent experiments are shown. C. CD8 or MX2 expressing U87-MG cells were challenged with GFP-encoding HIV-1-based vectors (containing wild-type CA, CAN74D, CAP90A or CA from SIVMAC) or an SIVMAC-based vector at an MOI of 0.25 as in Fig 4B. The percent of GFP-expressing cells was evaluated, and mean percentages of transduced cells for at least three independent experiments are shown. D. 293T cells were cotransfected with expression plasmids for GFP (Neg Ctrl), IFITM3, untagged and FLAG-tagged MX1 and MX2 (MX1-Fl and MX2-Fl), or the FLAG-tagged MX1 GTPase deficient mutants MX1K83A and MX1T103A along with an influenza A virus firefly luciferase minigenome plasmid and a renilla luciferase expression plasmid. At 24 h, cells were infected with influenza A virus A/Victoria/3/75 (H3N2) at a MOI of 2 and firefly and renilla luciferase activities were measured 18 h post-infection. Mean relative infection efficiencies for three independent experiments are shown. E. U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8, TRIMCyp, MX1, MX2 or the mutants MX1K83A, MX1T103A, MX2K131A. The cells were infected with 25 ng p24Gag of NL4-3/nef-IRES-renilla and infection efficiency was monitored at 48 h by measuring renilla activity. Mean relative infection efficiencies from three independent experiments are shown.
Comment in
- Viral infection: interfering with HIV infection.
Hofer U. Hofer U. Nat Rev Microbiol. 2013 Nov;11(11):742-3. doi: 10.1038/nrmicro3145. Epub 2013 Oct 8. Nat Rev Microbiol. 2013. PMID: 24100359 No abstract available. - Dynamins are forever: MxB inhibits HIV-1.
Haller O. Haller O. Cell Host Microbe. 2013 Oct 16;14(4):371-3. doi: 10.1016/j.chom.2013.10.002. Cell Host Microbe. 2013. PMID: 24139395
Similar articles
- Oligomerization Requirements for MX2-Mediated Suppression of HIV-1 Infection.
Dicks MD, Goujon C, Pollpeter D, Betancor G, Apolonia L, Bergeron JR, Malim MH. Dicks MD, et al. J Virol. 2015 Oct 7;90(1):22-32. doi: 10.1128/JVI.02247-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26446602 Free PMC article. - MX2 is an interferon-induced inhibitor of HIV-1 infection.
Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. Kane M, et al. Nature. 2013 Oct 24;502(7472):563-6. doi: 10.1038/nature12653. Epub 2013 Oct 13. Nature. 2013. PMID: 24121441 Free PMC article. - Equine Myxovirus Resistance Protein 2 Restricts Lentiviral Replication by Blocking Nuclear Uptake of Capsid Protein.
Ji S, Na L, Ren H, Wang Y, Wang X. Ji S, et al. J Virol. 2018 Aug 29;92(18):e00499-18. doi: 10.1128/JVI.00499-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29743377 Free PMC article. - Human MX2/MxB: a Potent Interferon-Induced Postentry Inhibitor of Herpesviruses and HIV-1.
Staeheli P, Haller O. Staeheli P, et al. J Virol. 2018 Nov 27;92(24):e00709-18. doi: 10.1128/JVI.00709-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258007 Free PMC article. Review. - Mx GTPases: dynamin-like antiviral machines of innate immunity.
Haller O, Staeheli P, Schwemmle M, Kochs G. Haller O, et al. Trends Microbiol. 2015 Mar;23(3):154-63. doi: 10.1016/j.tim.2014.12.003. Epub 2015 Jan 6. Trends Microbiol. 2015. PMID: 25572883 Review.
Cited by
- Cyclophilin A facilitates HIV-1 integration.
Padron A, Dwivedi R, Chakraborty R, Prakash P, Kim K, Shi J, Ahn J, Pandhare J, Luban J, Aiken C, Balasubramaniam M, Dash C. Padron A, et al. J Virol. 2024 Nov 19;98(11):e0094724. doi: 10.1128/jvi.00947-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480090 - Host cell glycosylation selects for infection with CCR5- versus CXCR4-tropic HIV-1.
Itell HL, Guenthoer J, Humes D, Baumgarten NE, Overbaugh J. Itell HL, et al. Nat Microbiol. 2024 Nov;9(11):2985-2996. doi: 10.1038/s41564-024-01806-7. Epub 2024 Oct 3. Nat Microbiol. 2024. PMID: 39363105 - Exploring HIV-1 Maturation: A New Frontier in Antiviral Development.
McGraw A, Hillmer G, Medehincu SM, Hikichi Y, Gagliardi S, Narayan K, Tibebe H, Marquez D, Mei Bose L, Keating A, Izumi C, Peese K, Joshi S, Krystal M, DeCicco-Skinner KL, Freed EO, Sardo L, Izumi T. McGraw A, et al. Viruses. 2024 Sep 6;16(9):1423. doi: 10.3390/v16091423. Viruses. 2024. PMID: 39339899 Free PMC article. Review. - Antiviral Mx proteins have an ancient origin and widespread distribution among eukaryotes.
Langley CA, Dietzen PA, Emerman M, Tenthorey JL, Malik HS. Langley CA, et al. bioRxiv [Preprint]. 2024 Aug 17:2024.08.06.606855. doi: 10.1101/2024.08.06.606855. bioRxiv. 2024. PMID: 39149278 Free PMC article. Preprint. - Human Immunodeficiency Virus-Induced Interferon-Stimulated Gene Expression Is Associated With Monocyte Activation and Predicts Viral Load.
van Pul L, van Dort KA, Girigorie AF, Maurer I, Harskamp AM, Kootstra NA. van Pul L, et al. Open Forum Infect Dis. 2024 Aug 5;11(8):ofae434. doi: 10.1093/ofid/ofae434. eCollection 2024 Aug. Open Forum Infect Dis. 2024. PMID: 39104769 Free PMC article.
References
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
- Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 098850/WT_/Wellcome Trust/United Kingdom
- G1000196/MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- DA033773/DA/NIDA NIH HHS/United States
- R01 DA033773/DA/NIDA NIH HHS/United States
- G1001081/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous