Piwi genes are dispensable for normal hematopoiesis in mice - PubMed (original) (raw)
Piwi genes are dispensable for normal hematopoiesis in mice
Mona J Nolde et al. PLoS One. 2013.
Abstract
Hematopoietic stem cells (HSC) must engage in a life-long balance between self-renewal and differentiation to sustain hematopoiesis. The highly conserved PIWI protein family regulates proliferative states of stem cells and their progeny in diverse organisms. A Human piwi gene (for clarity, the non-italicized "piwi" refers to the gene subfamily), HIWI (PIWIL1), is expressed in CD34⁺ stem/progenitor cells and transient expression of HIWI in a human leukemia cell line drastically reduces cell proliferation, implying the potential function of these proteins in hematopoiesis. Here, we report that one of the three piwi genes in mice, Miwi2 (Piwil4), is expressed in primitive hematopoetic cell types within the bone marrow. Mice with a global deletion of all three piwi genes, Miwi, Mili, and Miwi2, are able to maintain long-term hematopoiesis with no observable effect on the homeostatic HSC compartment in adult mice. The PIWI-deficient hematopoetic cells are capable of normal lineage reconstitution after competitive transplantation. We further show that the three piwi genes are dispensable during hematopoietic recovery after myeloablative stress by 5-FU. Collectively, our data suggest that the function of the piwi gene subfamily is not required for normal adult hematopoiesis.
Conflict of interest statement
Competing Interests: The authors declare no competing interests.
Figures
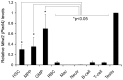
Figure 1. The mammalian Miwi2 gene is expressed in hematopoietic stem and progenitor cells.
Real-time PCR of FACS sorted young adult wild-type mouse bone marrow cells show expression of Miwi2 (Piwil4) at elevated levels in Lin− cell types, hematopoietic stem cells (HSC), multi-potent progenitors (MPP) and granulocyte-macrophage progenitors (GMP), but at low or undetectable levels in Lin+ cell types, red blood cells (RBC), macrophages (Mac), neutrophils (Neutr), B-cells, and T-cells. Quantitative PCR samples were normalized to Gapdh. Data is the average of triplicate samples from combined bone marrow from three mice. As indicated, the difference between the stem/progenitor cell types and the differentiated cell types is statistically significant. *P values: HSC vs: MPP = 0.667, GMP = 0.078, RBC = 0.012, Mac = 0.006, Neutr = 0.006, B-cell = 0.012, T-cell = 0.011; MPP vs: GMP = 0.146, RBC = 0.043, Mac = 0.028, Neutr = 0.029, B-cell = 0.044, T-cell = 0.042; GMP vs: RBC = 0.015, Mac = 0.012, Neutr = 0.012, B-cell = 0.015, T-cell = 0.015. P values calculated using two-tailed, equal variance t test.
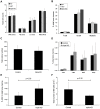
Figure 2. Deletion of all three piwi genes causes increased HSC frequency, but does not affect progenitor or committed cell type numbers.
Piwi-triple-knockout mature blood cell numbers are unchanged compared to control mice (WT, HET), as determined by (A) Complete Blood Cell Count (CBC) for white blood cells (WBC), neutrophils (NE), lymphocytes (LY), red blood cells (RBC) and (B) FACS analysis of committed bone marrow cell types (A: WT, n = 3; HET, n = 4; triple KO, n = 4; B: WT, n = 2; HET, n = 3, triple KO, n = 3). (C) Colony Forming Unit (CFU) assays show similar total myeloid/erythroid progenitor-derived colony numbers in control and triple Piwi mutant bone marrow. Colony assays used total bone marrow from 3 individual mice for each genotype. Bone marrow samples were plated in triplicate and total colonies were counted at days 7–10. Data shown as the average total colony count for each genotype. (D) In the triple Piwi mutant, the percentage of bone marrow progenitors (CMP, MEP and GMP) are only moderately increased compared to control mice progenitors. (E) HSC percentages are significantly increased in Piwi triple knockout bone marrow. (F) Total bone marrow cellularity of the triple knockout mice is almost two-fold reduced compared to control mice. (C-F: Control, n = 4; triple KO, n = 4). P values calculated using two-tailed, unequal variance Student's t test.
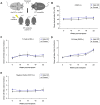
Figure 3. Piwi genes are not required for marrow HSC engraftment following irradiation.
(A) Illustration of competitive transplantation scheme whereby the triple piwi mutant or control donor CD45.2+ bone marrow is mixed equally with competitor CD45.1+ bone marrow and injected into a cohort of lethally irradiated CD45.1+ wild-type recipient mice. (B) Percentages of donor-derived (CD45.2+) cells from the triple knockout (blue circle) and control (black triangle) donors in the peripheral blood of lethally irradiated CD45.1+ recipient mice, measured up to five months (22 weeks) after transplant. (C-E) Ratio of donor/recipient (CD45.2+/CD45.2− ) derived cells within committed blood cell lineages, (C) T-cell (CD3+), (D) B-cell (B220+), and (E) myeloid cells (CD11b+). Data is the average of two independent experiments with 10 triple Piwi mutant injected recipients and 8 control injected recipients from each experiment. Each data point represents the mean ±SEM of 8–10 recipient mice, per donor genotype.
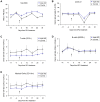
Figure 4. HSC myeloablative stress response is not affected by deletion of all piwi genes.
(A) White blood cells (WBC) are responsive to 5FU in both piwi triple knockout (blue circle) and control (black triangle) donor-derived cells. (B) Piwi triple knockout (blue circle) and control (black triangle) donor-derived cells (CD45.2+) show similar response kinetics following 5FU, measured up to 31 days after 5FU treatment. (C-E) Ratio of donor/recipient (CD45.2+/CD45.2− ) derived cells from Piwi triple knockout (blue circle) and control (black triangle) donors in mature cell populations of recipient peripheral blood. (C) T-cell (CD3+), (D) B-cell (B220+), and (E) myeloid cell (CD11b+) lineages. 5FU injection point is indicated as a black arrow on each graph. Data is the average of 10 triple Piwi mutant injected recipients and 9 control injected recipients. Each data point represents the mean ±SEM of 8–10 recipient mice, per donor genotype.
Similar articles
- Atypical protein kinase C (aPKCzeta and aPKClambda) is dispensable for mammalian hematopoietic stem cell activity and blood formation.
Sengupta A, Duran A, Ishikawa E, Florian MC, Dunn SK, Ficker AM, Leitges M, Geiger H, Diaz-Meco M, Moscat J, Cancelas JA. Sengupta A, et al. Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9957-62. doi: 10.1073/pnas.1103132108. Epub 2011 Jun 8. Proc Natl Acad Sci U S A. 2011. PMID: 21653884 Free PMC article. - Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo.
Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DI. Jacobs JE, et al. PLoS One. 2013 Dec 23;8(12):e82573. doi: 10.1371/journal.pone.0082573. eCollection 2013. PLoS One. 2013. PMID: 24376547 Free PMC article. - Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi.
Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Sharma AK, et al. Blood. 2001 Jan 15;97(2):426-34. doi: 10.1182/blood.v97.2.426. Blood. 2001. PMID: 11154219 - Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies.
Warr MR, Pietras EM, Passegué E. Warr MR, et al. Wiley Interdiscip Rev Syst Biol Med. 2011 Nov-Dec;3(6):681-701. doi: 10.1002/wsbm.145. Epub 2011 Mar 15. Wiley Interdiscip Rev Syst Biol Med. 2011. PMID: 21412991 Review. - Extravascular coagulation in hematopoietic stem and progenitor cell regulation.
Nguyen TS, Lapidot T, Ruf W. Nguyen TS, et al. Blood. 2018 Jul 12;132(2):123-131. doi: 10.1182/blood-2017-12-768986. Epub 2018 Jun 4. Blood. 2018. PMID: 29866813 Free PMC article. Review.
Cited by
- PIWIL1 gene polymorphism and pediatric acute lymphoblastic leukemia relapse susceptibility among Chinese children: a five-center case-control study.
Ding W, Wang D, Cai M, Yan Y, Liu S, Liu X, Luo A, Deng D, Liu X, Jiang H. Ding W, et al. Front Oncol. 2023 Nov 2;13:1203002. doi: 10.3389/fonc.2023.1203002. eCollection 2023. Front Oncol. 2023. PMID: 38023199 Free PMC article. - piRNA-like small RNAs mark extended 3'UTRs present in germ and somatic cells.
Yamtich J, Heo SJ, Dhahbi J, Martin DI, Boffelli D. Yamtich J, et al. BMC Genomics. 2015 Jun 16;16(1):462. doi: 10.1186/s12864-015-1662-6. BMC Genomics. 2015. PMID: 26076733 Free PMC article. - PIWI-mediated control of tissue-specific transposons is essential for somatic cell differentiation.
Li D, Taylor DH, van Wolfswinkel JC. Li D, et al. Cell Rep. 2021 Oct 5;37(1):109776. doi: 10.1016/j.celrep.2021.109776. Cell Rep. 2021. PMID: 34610311 Free PMC article. - PIWI proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells.
Cheng EC, Kang D, Wang Z, Lin H. Cheng EC, et al. PLoS One. 2014 Sep 19;9(9):e97821. doi: 10.1371/journal.pone.0097821. eCollection 2014. PLoS One. 2014. PMID: 25238487 Free PMC article. - Argonaute and Argonaute-Bound Small RNAs in Stem Cells.
Zhai L, Wang L, Teng F, Zhou L, Zhang W, Xiao J, Liu Y, Deng W. Zhai L, et al. Int J Mol Sci. 2016 Feb 4;17(2):208. doi: 10.3390/ijms17020208. Int J Mol Sci. 2016. PMID: 26861290 Free PMC article. Review.
References
- Civin CI, Strauss LC, Brovall C, Fackler MJ, Schwartz JF, et al. (1984) Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J Immunol 133: 157–165. - PubMed
- Krause DS, Ito T, Fackler MJ, Smith OM, Collector MI, et al. (1994) Characterization of murine CD34, a marker for hematopoietic progenitor and stem cells. Blood 84: 691–701. - PubMed
- Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, et al. (2001) CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97: 426–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases