Structure and nucleic acid binding activity of the nucleoporin Nup157 - PubMed (original) (raw)
Structure and nucleic acid binding activity of the nucleoporin Nup157
Hyuk-Soo Seo et al. Proc Natl Acad Sci U S A. 2013.
Abstract
At the center of the nuclear pore complex (NPC) is a uniquely versatile central transport channel. Structural analyses of distinct segments ("protomers") of the three "channel" nucleoporins yielded a model for how this channel is constructed. Its principal feature is a midplane ring that can undergo regulated diameter changes of as much as an estimated 30 nm. To better understand how a family of "adaptor" nucleoporins--concentrically surrounding this channel--might cushion these huge structural changes, we determined the crystal structure of one adaptor nucleoporin, Nup157. Here, we show that a recombinant Saccharomyces cerevisiae Nup157 protomer, representing two-thirds of Nup157 (residues 70-893), folds into a seven-bladed β-propeller followed by an α-helical domain, which together form a C-shaped architecture. Notably, the structure contains a large patch of positively charged residues, most of which are evolutionarily conserved. Consistent with this surface feature, we found that Nup157(70-893) binds to nucleic acids, although in a sequence-independent manner. Nevertheless, this interaction supports a previously reported role of Nup157, and its paralogue Nup170, in chromatin organization. Based on its nucleic acid binding capacity, we propose a dual location and function of Nup157. Finally, modeling the remaining C-terminal portion of Nup157 shows that it projects as a superhelical stack from the compact C-shaped portion of the molecule. The predicted four hinge regions indicate an intrinsic flexibility of Nup157, which could contribute to structural plasticity within the NPC.
Keywords: DNA-binding protein; RNA-binding protein; X-ray crystallography; gene gating.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure of the S. cerevisiae Nup15770–893. (A) Schematic representation of the Nup157 domain architecture. The boundaries for each domain are color-coded and marked with residue numbers. Starting from the N terminus: the unstructured fragment (gray) denoted by “U”, β-propeller domain (orange), α-helical domain (yellow), and a predicted α-helical region (gray) are indicated. Positions of the N-terminal α-helical extension (α1) and five α-helical insertions (α2–α6) in the propeller domain are indicated: α1/α5/α6, in blue, create an interface between the two domains in Nup157, whereas α2 to α4, in green, are found on the side of the propeller. The bar above the domain architecture corresponds to the crystallized fragment (residues 70–893). (B) Structure of Nup15770–893 in ribbon representation, colored as in A. (Right) A 90°-rotated view. (C) Ribbon representation of the Nup157 β-propeller domain. Seven blades of the β-propeller core (orange), the α-helical insertions (blue and green) are indicated. (Right) Schematic representation of the Nup15770–893 β-propeller domain and locations of its α-helical elements. Dotted gray lines indicate disordered regions. The double Velcro-type closure is highlighted by asterisks.
Fig. 2.
Interface between the Nup15770–893 domains. (A) Space-filling model of Nup15770–893 (Center) and dissection of the buried interface between the β-propeller (orange; Right) and α-helical domain (yellow; Left). The α1 extension and α2 to α6 insertions are color-coded as in Fig. 1. Residues involved in forming the interface between the two domains are in magenta and contoured using a black line. (B) Surface representation of Nup15770–893 is shown in the same view as in A. Conserved residues from multiple sequence alignment (
Fig. S1
) are mapped onto the surface and shaded in a color gradient from light yellow to dark red (40–100% sequence conservation). A magnified view in ribbon representation of the interdomain interface highlighting hydrophilic (C) and hydrophobic (D) interactions formed by α1/α5/α6 (blue) in the β-propeller and α9/α11 (yellow) in the α-helical domain.
Fig. 3.
Surface properties of Nup15770–893. (A) Conserved residues from multiple sequence alignment (
Fig. S1
) are mapped onto the surface and shaded in a color gradient from light yellow to red (40–100% sequence conservation). (B) Electrostatic potential surface of Nup15770–893 is depicted in a gradient from red (−5 kT/e) to blue (+5 kT/e) as indicated (Right). The orientation of surface representations is identical in each column. The three most sequence conserved surface patches (areas 1–3) are encircled by a black line.
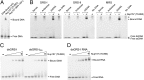
Fig. 4.
Nup157 directly interacts with nucleic acids in vitro. (A) EMSA was used to probe the interaction of Nup15770–893 and Nup157894–1391 with DNA. Protein samples were mixed with a DNA library (refer to
Table S2
) at a 4:1 molar ratio and analyzed by native PAGE. Free and bound DNAs were visualized by SYBR Gold staining as indicated by black arrows. (B) Three INO1 zip codes (GRS-I and –II and MRS) were tested for Nup157 binding. Eightfold molar excess of Nup15770–893 over ssDNA (sense or antisense 21-mer DNA) or dsDNA was used for EMSA. Background staining of Nup15770–893 is marked by an asterisk. (C) Titration of Nup15770–893 to GRS-I and the reversed GRS-I (GRS-IRev). Both GRS elements were preincubated with increasing amounts of Nup15770–893 at 0-, 0.3-, 1-, 2-, 5-, 10-, and 20-fold molar excess and analyzed by EMSA. (D) Titration of Nup15770–893 to the RNA equivalent of the GRS-I element as in C.
Fig. 5.
A structural model of Nup157. (A) A schematic representation of the Nup157 domain architecture is shown as in Fig 1_A_. The dotted line above the α-helical region corresponds to the modeled C-terminal remainder of Nup157 (residues 894–1391). (B) Structural comparison between Nup170 (residues 979–1502; Protein Data Bank ID code 3I5P) in cyan and Nup157 (homology model) in gray. The structures align with 1.2 Å Cα rmsd. (C) A composite model of Nup157 (residues 70–1391) is colored as in A. The N-terminal, 69-residue fragment of Nup157 is predicted to be disordered and was therefore omitted from the 3D structure prediction. Four hinge residues (hinge 1-T108, hinge 2-R615, hinge 3-R1053, hinge 4-L1241) predicted by elastic network models are indicated by red arrows.
Similar articles
- Reconstitution of Nup157 and Nup145N into the Nup84 complex.
Lutzmann M, Kunze R, Stangl K, Stelter P, Tóth KF, Böttcher B, Hurt E. Lutzmann M, et al. J Biol Chem. 2005 May 6;280(18):18442-51. doi: 10.1074/jbc.M412787200. Epub 2005 Mar 1. J Biol Chem. 2005. PMID: 15741174 - Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins.
Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A. Stuwe T, et al. Proc Natl Acad Sci U S A. 2014 Feb 18;111(7):2530-5. doi: 10.1073/pnas.1311081111. Epub 2014 Feb 6. Proc Natl Acad Sci U S A. 2014. PMID: 24505056 Free PMC article. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review.
Cited by
- Trafficking and/or division: Distinct roles of nucleoporins based on their location within the nuclear pore complex.
Hegedűsová E, Maršalová V, Kulkarni S, Paris Z. Hegedűsová E, et al. RNA Biol. 2022;19(1):650-661. doi: 10.1080/15476286.2022.2067711. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35491934 Free PMC article. - Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - Nucleoporin insufficiency disrupts a pluripotent regulatory circuit in a pro-arrhythmogenic stem cell line.
Preston CC, Storm EC, Burdine RD, Bradley TA, Uttecht AD, Faustino RS. Preston CC, et al. Sci Rep. 2019 Sep 3;9(1):12691. doi: 10.1038/s41598-019-49147-4. Sci Rep. 2019. PMID: 31481660 Free PMC article. - Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98).
Quan B, Seo HS, Blobel G, Ren Y. Quan B, et al. Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9127-32. doi: 10.1073/pnas.1409076111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927547 Free PMC article. - The Nuclear Pore Complex as a Flexible and Dynamic Gate.
Knockenhauer KE, Schwartz TU. Knockenhauer KE, et al. Cell. 2016 Mar 10;164(6):1162-1171. doi: 10.1016/j.cell.2016.01.034. Cell. 2016. PMID: 26967283 Free PMC article. Review.
References
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
- Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315(5819):1729–1732. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases