Reliable identification of genomic variants from RNA-seq data - PubMed (original) (raw)
Reliable identification of genomic variants from RNA-seq data
Robert Piskol et al. Am J Hum Genet. 2013.
Abstract
Identifying genomic variation is a crucial step for unraveling the relationship between genotype and phenotype and can yield important insights into human diseases. Prevailing methods rely on cost-intensive whole-genome sequencing (WGS) or whole-exome sequencing (WES) approaches while the identification of genomic variants from often existing RNA sequencing (RNA-seq) data remains a challenge because of the intrinsic complexity in the transcriptome. Here, we present a highly accurate approach termed SNPiR to identify SNPs in RNA-seq data. We applied SNPiR to RNA-seq data of samples for which WGS and WES data are also available and achieved high specificity and sensitivity. Of the SNPs called from the RNA-seq data, >98% were also identified by WGS or WES. Over 70% of all expressed coding variants were identified from RNA-seq, and comparable numbers of exonic variants were identified in RNA-seq and WES. Despite our method's limitation in detecting variants in expressed regions only, our results demonstrate that SNPiR outperforms current state-of-the-art approaches for variant detection from RNA-seq data and offers a cost-effective and reliable alternative for SNP discovery.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
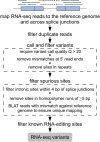
Figure 1
A Computational Framework for the Identification of SNPs from Transcriptome Data Shown are RNA-seq reads mapped to the human reference genome (blue lines) and all regions spanning known splice junctions (yellow lines separated by dashes). Subsequent variant calling used GATK and filtering to remove spurious sites, generating a high-confidence set of SNVs.
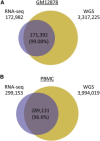
Figure 2
Comparison of SNPs Identified via RNA-Seq and WGS of GM12878 Cells and PBMCs SNPiR achieved high precision for both GM12878 (A) and PBMC (B) data sets, given that most of the RNA-seq variants were also identified by WGS of the same subject. Numbers in parentheses give the percentage of RNA-seq variants found in WGS.
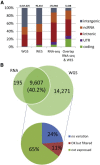
Figure 3
Characteristics of SNPs Identified from RNA-Seq Data of GM12878 Cells (A) The composition of genomic regions for variants in WGS, WES, and RNA-seq suggests a high enrichment of RNA-seq variants in functionally important regions. Sites present in RNA-seq and WES occurred substantially more often in coding exons. (B) Overlap in coding variants detected from RNA-seq and WGS. Of all coding variants, 40.2% were found by RNA-seq. The majority of the remaining sites were not detected as a result of the lack of expression. “No variation” indicates that the position was homozygous in RNA, “OK but filtered” indicates that the position was heterozygous but was removed by one of our filtering steps, and “not expressed” indicates that the position was not covered by RNA-seq reads.
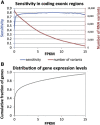
Figure 4
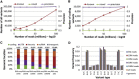
High Sensitivity of SNPiR Variant Calling in Coding Regions of Expressed Genes of GM12878 Cells (A) Sensitivity and number of detected variants called from RNA-seq data in dependence of the minimum gene expression (in FPKM). (B) Cumulative distribution of expression levels (in FPKM) for all reference genes.
Figure 5
Subsampling of RNA-Seq Reads Subsamplings of 5, 10, 20, 50, and 100 million reads were generated from the total set of 499 million GM12878 RNA-seq reads. We compared (A) the number of discovered variants, (B) the number of variants in coding regions, and (C) the genomic location of variants between the random samplings and the complete set RNA-seq reads, as well as (D) the mutational profile of known RNA-seq variants and genomic variants. In (A) and (B), “known” variants denote all variant sites that were discovered from RNA-seq and were either confirmed through WGS or present in dbSNP. Conversely, “novel” denotes all variants that were previously not found from WGS or dbSNP. The total amounts of novel variants per sample size are shown as small numbers above the data series.
Figure 6
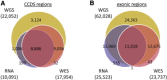
Comparison of Genomic Variants Identified in CCDS and Exonic Regions by WGS, WES, or RNA-Seq An equal number of reads (94.1 million) of RNA-seq and WES data was used for fair comparison of variants identified in CCDS regions (A) and exonic regions (B).≥
Figure 7
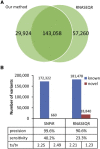
Comparison of SNPiR with RNASEQR Overlap between the sites detected by SNPiR and RNASEQR on the same RNA-seq data set for GM12878 cells (A) and the number of known and novel variants discovered by SNPiR and RNASEQR, the precision and sensitivity of variant calling, and the ts/tv ratio for each category (B). Precision was calculated as the fraction of RNA-seq variants either supported by WGS or present in dbSNP. Sensitivity was determined as the fraction of WGS variants both found in coding regions and discovered in the RNA-seq data.
Similar articles
- Variant detection sensitivity and biases in whole genome and exome sequencing.
Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Meynert AM, et al. BMC Bioinformatics. 2014 Jul 19;15(1):247. doi: 10.1186/1471-2105-15-247. BMC Bioinformatics. 2014. PMID: 25038816 Free PMC article. - Variant analysis pipeline for accurate detection of genomic variants from transcriptome sequencing data.
Adetunji MO, Lamont SJ, Abasht B, Schmidt CJ. Adetunji MO, et al. PLoS One. 2019 Sep 23;14(9):e0216838. doi: 10.1371/journal.pone.0216838. eCollection 2019. PLoS One. 2019. PMID: 31545812 Free PMC article. - Inconsistency and features of single nucleotide variants detected in whole exome sequencing versus transcriptome sequencing: A case study in lung cancer.
O'Brien TD, Jia P, Xia J, Saxena U, Jin H, Vuong H, Kim P, Wang Q, Aryee MJ, Mino-Kenudson M, Engelman JA, Le LP, Iafrate AJ, Heist RS, Pao W, Zhao Z. O'Brien TD, et al. Methods. 2015 Jul 15;83:118-27. doi: 10.1016/j.ymeth.2015.04.016. Epub 2015 Apr 23. Methods. 2015. PMID: 25913717 Free PMC article. - Single-nucleotide variants in human RNA: RNA editing and beyond.
Guo Y, Yu H, Samuels DC, Yue W, Ness S, Zhao YY. Guo Y, et al. Brief Funct Genomics. 2019 Feb 14;18(1):30-39. doi: 10.1093/bfgp/ely032. Brief Funct Genomics. 2019. PMID: 30312373 Free PMC article. Review. - Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies.
Chou J, Ohsumi TK, Geha RS. Chou J, et al. Curr Opin Allergy Clin Immunol. 2012 Dec;12(6):623-8. doi: 10.1097/ACI.0b013e3283588ca6. Curr Opin Allergy Clin Immunol. 2012. PMID: 23095910 Review.
Cited by
- Practicability of detecting somatic point mutation from RNA high throughput sequencing data.
Sheng Q, Zhao S, Li CI, Shyr Y, Guo Y. Sheng Q, et al. Genomics. 2016 May;107(5):163-9. doi: 10.1016/j.ygeno.2016.03.006. Epub 2016 Apr 2. Genomics. 2016. PMID: 27046520 Free PMC article. - SNP calling from RNA-seq data without a reference genome: identification, quantification, differential analysis and impact on the protein sequence.
Lopez-Maestre H, Brinza L, Marchet C, Kielbassa J, Bastien S, Boutigny M, Monnin D, Filali AE, Carareto CM, Vieira C, Picard F, Kremer N, Vavre F, Sagot MF, Lacroix V. Lopez-Maestre H, et al. Nucleic Acids Res. 2016 Nov 2;44(19):e148. doi: 10.1093/nar/gkw655. Epub 2016 Jul 25. Nucleic Acids Res. 2016. PMID: 27458203 Free PMC article. - Indel detection from RNA-seq data: tool evaluation and strategies for accurate detection of actionable mutations.
Sun Z, Bhagwate A, Prodduturi N, Yang P, Kocher JA. Sun Z, et al. Brief Bioinform. 2017 Nov 1;18(6):973-983. doi: 10.1093/bib/bbw069. Brief Bioinform. 2017. PMID: 27473065 Free PMC article. - Targeted RNA Sequencing Assay to Characterize Gene Expression and Genomic Alterations.
Martin DP, Miya J, Reeser JW, Roychowdhury S. Martin DP, et al. J Vis Exp. 2016 Aug 4;(114):54090. doi: 10.3791/54090. J Vis Exp. 2016. PMID: 27585245 Free PMC article. - Practical aspects of NGS-based pathways analysis for personalized cancer science and medicine.
Kotelnikova EA, Pyatnitskiy M, Paleeva A, Kremenetskaya O, Vinogradov D. Kotelnikova EA, et al. Oncotarget. 2016 Aug 9;7(32):52493-52516. doi: 10.18632/oncotarget.9370. Oncotarget. 2016. PMID: 27191992 Free PMC article. Review.
References
- Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources