Differential abundance analysis for microbial marker-gene surveys - PubMed (original) (raw)
Differential abundance analysis for microbial marker-gene surveys
Joseph N Paulson et al. Nat Methods. 2013 Dec.
Abstract
We introduce a methodology to assess differential abundance in sparse high-throughput microbial marker-gene survey data. Our approach, implemented in the metagenomeSeq Bioconductor package, relies on a novel normalization technique and a statistical model that accounts for undersampling-a common feature of large-scale marker-gene studies. Using simulated data and several published microbiota data sets, we show that metagenomeSeq outperforms the tools currently used in this field.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
Figure 1. Clustering analysis is improved substantially by CSS normalization
We plot the first two principal coordinates in a multi-dimensional scaling analysis of mouse stool data normalized by (A) CSS, (B) DESeq size factors, (C) trimmed mean of M-values, and (D) total-sum. Colors indicate clinical phenotype (diet). CSS normalization data successfully separates samples by diet while controlling within-group variability. (E) Class posterior probability log-ratio for Western diet obtained from linear discriminant analysis (LDA). Each box corresponds to the distribution of leave-one-out posterior probability of assignment to the “Western” cluster across normalization methods (whiskers indicate 1.5 times inter-quartile range). Samples were best distinguished by phenotypic similarity using CSS normalization.
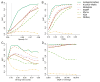
Figure 2. Simulation results indicate that metagenomeSeq has greater sensitivity and specificity in a variety of settings
We use area under the receiver operating characteristic curve (AUC) to compare Metastats, Xipe, Kruskal-Wallis test as used in Lefse, a non-zero inflated log-normal model, edgeR and DESeq. (A) AUC as dataset sparsity decreases. MetagenomeSeq achieves larger AUC values than any other method in datasets with high sparsity (vertical dashed line represents the least sparse metagenomic dataset). (B) AUC as the effect-size between two conditions increases. Both metagenomeSeq and Lefse are better at detecting features with small effect size. (C) AUC as the variability in depth of sequencing increases. MetagenomeSeq and Kruskal-Wallis are robust to high variability in sequencing depth. (D) AUC as average sequencing depth increases. All models (except the non-zero inflated log-normal model and XIPE) perform similarly well at sufficient depth of coverage.
Comment in
- A fair comparison.
Costea PI, Zeller G, Sunagawa S, Bork P. Costea PI, et al. Nat Methods. 2014 Apr;11(4):359. doi: 10.1038/nmeth.2897. Nat Methods. 2014. PMID: 24681719 No abstract available.
Similar articles
- Metagenomic species profiling using universal phylogenetic marker genes.
Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, Rasmussen S, Brunak S, Pedersen O, Guarner F, de Vos WM, Wang J, Li J, Doré J, Ehrlich SD, Stamatakis A, Bork P. Sunagawa S, et al. Nat Methods. 2013 Dec;10(12):1196-9. doi: 10.1038/nmeth.2693. Epub 2013 Oct 20. Nat Methods. 2013. PMID: 24141494 - Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data.
Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Davis NM, et al. Microbiome. 2018 Dec 17;6(1):226. doi: 10.1186/s40168-018-0605-2. Microbiome. 2018. PMID: 30558668 Free PMC article. - A framework for assessing 16S rRNA marker-gene survey data analysis methods using mixtures.
Olson ND, Kumar MS, Li S, Braccia DJ, Hao S, Timp W, Salit ML, Stine OC, Bravo HC. Olson ND, et al. Microbiome. 2020 Mar 13;8(1):35. doi: 10.1186/s40168-020-00812-1. Microbiome. 2020. PMID: 32169095 Free PMC article. - Compositional data analysis of the microbiome: fundamentals, tools, and challenges.
Tsilimigras MC, Fodor AA. Tsilimigras MC, et al. Ann Epidemiol. 2016 May;26(5):330-5. doi: 10.1016/j.annepidem.2016.03.002. Epub 2016 Mar 31. Ann Epidemiol. 2016. PMID: 27255738 Review. - Meta'omic analytic techniques for studying the intestinal microbiome.
Morgan XC, Huttenhower C. Morgan XC, et al. Gastroenterology. 2014 May;146(6):1437-1448.e1. doi: 10.1053/j.gastro.2014.01.049. Epub 2014 Jan 28. Gastroenterology. 2014. PMID: 24486053 Review.
Cited by
- Timing matters: age-dependent impacts of the social environment and host selection on the avian gut microbiota.
Maraci Ö, Antonatou-Papaioannou A, Jünemann S, Engel K, Castillo-Gutiérrez O, Busche T, Kalinowski J, Caspers BA. Maraci Ö, et al. Microbiome. 2022 Nov 26;10(1):202. doi: 10.1186/s40168-022-01401-0. Microbiome. 2022. PMID: 36434663 Free PMC article. - The Effect of Plant Geographical Location and Developmental Stage on Root-Associated Microbiomes of Gymnadenia conopsea.
Lin M, Xiong H, Xiang X, Zhou Z, Liang L, Mei Z. Lin M, et al. Front Microbiol. 2020 Jun 18;11:1257. doi: 10.3389/fmicb.2020.01257. eCollection 2020. Front Microbiol. 2020. PMID: 32625183 Free PMC article. - An Overview of Bioinformatics Tools for DNA Meta-Barcoding Analysis of Microbial Communities of Bioaerosols: Digest for Microbiologists.
Mbareche H, Dumont-Leblond N, Bilodeau GJ, Duchaine C. Mbareche H, et al. Life (Basel). 2020 Sep 8;10(9):185. doi: 10.3390/life10090185. Life (Basel). 2020. PMID: 32911871 Free PMC article. Review. - Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses.
Pérez-Cobas AE, Gomez-Valero L, Buchrieser C. Pérez-Cobas AE, et al. Microb Genom. 2020 Aug;6(8):mgen000409. doi: 10.1099/mgen.0.000409. Epub 2020 Jul 24. Microb Genom. 2020. PMID: 32706331 Free PMC article. Review. - Vitamin D and allergic airway disease shape the murine lung microbiome in a sex-specific manner.
Roggenbuck M, Anderson D, Barfod KK, Feelisch M, Geldenhuys S, Sørensen SJ, Weeden CE, Hart PH, Gorman S. Roggenbuck M, et al. Respir Res. 2016 Sep 21;17(1):116. doi: 10.1186/s12931-016-0435-3. Respir Res. 2016. PMID: 27655266 Free PMC article.
References
- Kåhrström CT. Microbiome: Gut microbiome as a marker for diabetes. Nature Reviews Microbiology. 2012;10
- Harris JK, Wagner BD. Bacterial identification and analytic challenges in clinical microbiome studies. J Allergy Clin Immunol. 2012;129:441–442. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources