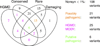Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes - PubMed (original) (raw)
. 2013 Nov;45(11):1380-5.
doi: 10.1038/ng.2794. Epub 2013 Oct 6.
Nicola L Beer, Alexander G Bick, Vineeta Agarwala, Janne Molnes, Namrata Gupta, Noël P Burtt, Jose C Florez, James B Meigs, Herman Taylor, Valeriya Lyssenko, Henrik Irgens, Ervin Fox, Frank Burslem, Stefan Johansson, M Julia Brosnan, Jeff K Trimmer, Christopher Newton-Cheh, Tiinamaija Tuomi, Anders Molven, James G Wilson, Christopher J O'Donnell, Sekar Kathiresan, Joel N Hirschhorn, Pål R Njølstad, Tim Rolph, J G Seidman, Stacey Gabriel, David R Cox, Christine E Seidman, Leif Groop, David Altshuler
Affiliations
- PMID: 24097065
- PMCID: PMC4051627
- DOI: 10.1038/ng.2794
Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes
Jason Flannick et al. Nat Genet. 2013 Nov.
Abstract
Genome sequencing can identify individuals in the general population who harbor rare coding variants in genes for Mendelian disorders and who may consequently have increased disease risk. Previous studies of rare variants in phenotypically extreme individuals display ascertainment bias and may demonstrate inflated effect-size estimates. We sequenced seven genes for maturity-onset diabetes of the young (MODY) in well-phenotyped population samples (n = 4,003). We filtered rare variants according to two prediction criteria for disease-causing mutations: reported previously in MODY or satisfying stringent de novo thresholds (rare, conserved and protein damaging). Approximately 1.5% and 0.5% of randomly selected individuals from the Framingham and Jackson Heart Studies, respectively, carry variants from these two classes. However, the vast majority of carriers remain euglycemic through middle age. Accurate estimates of variant effect sizes from population-based sequencing are needed to avoid falsely predicting a substantial fraction of individuals as being at risk for MODY or other Mendelian diseases.
Figures
Figure 1. Description of low frequency non-synonymous variants
Shown is a summary of all low frequency (MAF<1%) non-synonymous variants identified in the three cohorts (Supplementary Tables 1–3 show lists of all variants, not simply those of low frequency). MAF is calculated as the maximum frequency across the three cohorts. Shown also is the number of variants fitting each annotation (HGMD, conserved, rare, and damaging) and each variant class: low frequency non-synonymous (black, magenta, orange, or green), possibly pathogenic (orange or green), HGMD MODY (magenta or green), and putative pathogenic (green). Twelve low frequency variants fit no annotations. Low frequency non-synonymous is abbreviated to ‘Nonsyn <1%’.
Figure 2. Phenotypic impact of variants in unselected cohorts
Shown is the fraction of variant carriers in the two unselected cohorts with (a) diabetes and (b) IFG or diabetes. Separate fractions are given for each of the four defined variant classes, with the fraction of non-carriers with each phenotype shown as reference. Error bars reflect 68% confidence intervals in the estimated fractions and are computed via the Clopper-Pearson method. The number of analyzed samples is given in Supplementary Table 6. Fewer individuals had FPG measurements than diabetes measurements; thus the number of analyzed individuals for these two phenotypes differs. ‘Nonsyn <1%’ is low frequency non-synonymous, ‘Pos. Pathogenic’ is possibly pathogenic, and ‘Put. Pathogenic’ is putative pathogenic.
Figure 3. Phenotypes of GCK and HNF1A variant carriers
(a) Shown are FPG values for GCK-variant carriers in the FHS cohort (left) and JHS cohort (right). Three dashed lines correspond to defined FPG thresholds: top (solid) line represents diabetes (126 mg/dL), middle (dashed) line IFG (100 mg/dL), bottom (dotted) line _GCK_-MODY (99mg/dL). A histogram and box plot representing FPG levels in the non-diabetic population (computed separately for each cohort) is shown for comparison. Two GCK variant carriers were on medication for diabetes and are thus excluded from the plot. Tabular form of these results (including the two carriers with diabetes) is shown in Supplementary Table 10. (b) The scatter plot shows fasting-2hr post-OGTT plasma glucose increment (y-axis) and FPG (x-axis) for each _HNF1A_-variant carrier in the FHS cohort (OGTT information was unavailable for the JHS cohort). Histograms showing FPG and fasting-2hr post-OGTT plasma glucose increments for individuals in the FHS cohort without diabetes are shown on the left and below the scatter plot respectively. FPG values for individuals receiving treatment for diabetes were omitted from the plot. The vertical and horizontal dashed lines represent the IFG threshold (100mg/dL) and a plasma glucose increment consistent with _HNF1A_-MODY/beta-cell dysfunction (≥90mg/dL) respectively. Points are colored corresponding to the annotation class of the variant; for variants that belong to multiple classes, colors are chosen according to the following precedence: putative pathogenic, HGMD MODY, possibly pathogenic, low frequency non-synonymous.
Similar articles
- Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes.
Winckler W, Weedon MN, Graham RR, McCarroll SA, Purcell S, Almgren P, Tuomi T, Gaudet D, Boström KB, Walker M, Hitman G, Hattersley AT, McCarthy MI, Ardlie KG, Hirschhorn JN, Daly MJ, Frayling TM, Groop L, Altshuler D. Winckler W, et al. Diabetes. 2007 Mar;56(3):685-93. doi: 10.2337/db06-0202. Diabetes. 2007. PMID: 17327436 - Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry.
Johansson BB, Irgens HU, Molnes J, Sztromwasser P, Aukrust I, Juliusson PB, Søvik O, Levy S, Skrivarhaug T, Joner G, Molven A, Johansson S, Njølstad PR. Johansson BB, et al. Diabetologia. 2017 Apr;60(4):625-635. doi: 10.1007/s00125-016-4167-1. Epub 2016 Dec 2. Diabetologia. 2017. PMID: 27913849 - Causal variants in Maturity Onset Diabetes of the Young (MODY) - A systematic review.
Rafique I, Mir A, Saqib MAN, Naeem M, Marchand L, Polychronakos C. Rafique I, et al. BMC Endocr Disord. 2021 Nov 11;21(1):223. doi: 10.1186/s12902-021-00891-7. BMC Endocr Disord. 2021. PMID: 34763692 Free PMC article. - Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns.
Bonnycastle LL, Willer CJ, Conneely KN, Jackson AU, Burrill CP, Watanabe RM, Chines PS, Narisu N, Scott LJ, Enloe ST, Swift AJ, Duren WL, Stringham HM, Erdos MR, Riebow NL, Buchanan TA, Valle TT, Tuomilehto J, Bergman RN, Mohlke KL, Boehnke M, Collins FS. Bonnycastle LL, et al. Diabetes. 2006 Sep;55(9):2534-40. doi: 10.2337/db06-0178. Diabetes. 2006. PMID: 16936201 - [MODY].
Sasaki A, Horikawa Y, Takeda J. Sasaki A, et al. Nihon Rinsho. 2006 Sep 28;Suppl 3:54-8. Nihon Rinsho. 2006. PMID: 17022499 Review. Japanese. No abstract available.
Cited by
- Targeted sequencing identifies novel variants in common and rare MODY genes.
de Santana LS, Caetano LA, Costa-Riquetto AD, Franco PC, Dotto RP, Reis AF, Weinert LS, Silveiro SP, Vendramini MF, do Prado FA, Abrahão GCP, de Almeida AGFP, Tavares MDGR, Gonçalves WRB, Santomauro Junior AC, Halpern B, Jorge AAL, Nery M, Teles MG. de Santana LS, et al. Mol Genet Genomic Med. 2019 Dec;7(12):e962. doi: 10.1002/mgg3.962. Epub 2019 Oct 8. Mol Genet Genomic Med. 2019. PMID: 31595705 Free PMC article. - Disease variants in genomes of 44 centenarians.
Freudenberg-Hua Y, Freudenberg J, Vacic V, Abhyankar A, Emde AK, Ben-Avraham D, Barzilai N, Oschwald D, Christen E, Koppel J, Greenwald B, Darnell RB, Germer S, Atzmon G, Davies P. Freudenberg-Hua Y, et al. Mol Genet Genomic Med. 2014 Sep;2(5):438-50. doi: 10.1002/mgg3.86. Epub 2014 Jun 15. Mol Genet Genomic Med. 2014. PMID: 25333069 Free PMC article. - Guidelines for investigating causality of sequence variants in human disease.
MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. MacArthur DG, et al. Nature. 2014 Apr 24;508(7497):469-76. doi: 10.1038/nature13127. Nature. 2014. PMID: 24759409 Free PMC article. - An intermediate-effect size variant in UMOD confers risk for chronic kidney disease.
Olinger E, Schaeffer C, Kidd K, Elhassan EAE, Cheng Y, Dufour I, Schiano G, Mabillard H, Pasqualetto E, Hofmann P, Fuster DG, Kistler AD, Wilson IJ, Kmoch S, Raymond L, Robert T; Genomics England Research Consortium; Eckardt KU, Bleyer AJ Sr, Köttgen A, Conlon PJ, Wiesener M, Sayer JA, Rampoldi L, Devuyst O. Olinger E, et al. Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2114734119. doi: 10.1073/pnas.2114734119. Epub 2022 Aug 10. Proc Natl Acad Sci U S A. 2022. PMID: 35947615 Free PMC article. - Emerging roles of rare and low-frequency genetic variants in type 1 diabetes mellitus.
Pang H, Xia Y, Luo S, Huang G, Li X, Xie Z, Zhou Z. Pang H, et al. J Med Genet. 2021 May;58(5):289-296. doi: 10.1136/jmedgenet-2020-107350. Epub 2021 Mar 22. J Med Genet. 2021. PMID: 33753534 Free PMC article. Review.
References
- Collins FS. Shattuck lecture--medical and societal consequences of the Human Genome Project. The New England Journal of Medicine. 1999;341:28–37. - PubMed
- Collins FS. Genetics: an explosion of knowledge is transforming clinical practice. Geriatrics. 1999;54:41–47. quiz 48. - PubMed
- Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. - PubMed
- Hall Y. Coming Soon: Your Personal DNA Map? National Geographic News. 2006
- Duncan DE. On a Mission to Sequence the Genomes of 100,000 People. New York Times. 2010
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- U54 HG003067/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 HL080494/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- N01 HC095170/HL/NHLBI NIH HHS/United States
- K24 DK080140/DK/NIDDK NIH HHS/United States
- 6R01-NS 17950/NS/NINDS NIH HHS/United States
- N01-HC-95172/HC/NHLBI NIH HHS/United States
- T32GM007753/GM/NIGMS NIH HHS/United States
- N01 HC095171/HL/NHLBI NIH HHS/United States
- R01 DK078616/DK/NIDDK NIH HHS/United States
- N02-HL-6-4278/HL/NHLBI NIH HHS/United States
- 5-T32-GM007748-33/GM/NIGMS NIH HHS/United States
- N01-HC-25195/HC/NHLBI NIH HHS/United States
- N01 HC095172/HL/NHLBI NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- R01 2R01HL080494/HL/NHLBI NIH HHS/United States
- N01-HC-95171/HC/NHLBI NIH HHS/United States
- R01 HL084553/HL/NHLBI NIH HHS/United States
- N01-HC-95170/HC/NHLBI NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical