Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins - PubMed (original) (raw)
. 2013 Dec;31(12):1137-42.
doi: 10.1038/nbt.2726. Epub 2013 Oct 9.
James F Angstman, Marcy E Richardson, Samantha J Linder, Vincent M Cascio, Shengdar Q Tsai, Quan H Ho, Jeffry D Sander, Deepak Reyon, Bradley E Bernstein, Joseph F Costello, Miles F Wilkinson, J Keith Joung
Affiliations
- PMID: 24108092
- PMCID: PMC3858462
- DOI: 10.1038/nbt.2726
Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins
Morgan L Maeder et al. Nat Biotechnol. 2013 Dec.
Abstract
Genome-wide studies have defined cell type-specific patterns of DNA methylation that are important for regulating gene expression in both normal development and disease. However, determining the functional significance of specific methylation events remains challenging, owing to the lack of methods for removing such modifications in a targeted manner. Here we describe an approach for efficient targeted demethylation of specific CpGs in human cells using fusions of engineered transcription activator-like effector (TALE) repeat arrays and the TET1 hydroxylase catalytic domain. Using these TALE-TET1 fusions, we demonstrate that modification of critical methylated promoter CpG positions can lead to substantial increases in the expression of endogenous human genes. Our results delineate a strategy for understanding the functional significance of specific CpG methylation marks in the context of endogenous gene loci and validate programmable DNA demethylation reagents with potential utility for research and therapeutic applications.
Conflict of interest statement
Conflict of Interest Statement
M.L.M., J.F.A., and J.K.J. have filed a provisional patent application covering the TALE-TET fusion proteins. J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Figures
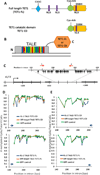
Figure 1. TALE-TET1 Fusion Proteins Induce Targeted Demethylation in Human Cells
(A) Schematic illustrating the predicted domain architecture of the full-length TET1 protein (TET1 FL) and the catalytic domain of the TET1 protein (TET1 CD). CXXC = CXXC-type zinc-binding domain, NLS = nuclear localization signal, Cys-rich = cysteine-rich region, DSBH = double-stranded β helix domain (DSBH). (B) Schematic illustrating general structure of fusions between a TALE repeat array DNA-binding domain and either TET1 FL or TET1 CD. (C) Schematic illustrating the human KLF4 locus with CpGs indicated with black arrows. Red arrows indicate the location and direction (5’ to 3’) of TALE-TET1 fusion protein binding sites. Numbering on the bottom line indicates position on the DNA relative to the start site of transcription (right-angle arrow) and numbering on the top line indicates position relative to the beginning of intron 2/3. (D) Demethylation activities of KL-1 TALE-TET1 CD (top) and KL-1 TALE-TET1 FL (bottom) fusion proteins in human K562 cells. Graphs show the fraction of CpGs methylated (y-axis) for different positions along the length of the second KLF4 intron (x-axis, numbered relative to the start of the intron). Blue arrow indicates the location and direction (5’ to 3’) of TALE target binding site. Each point represents the mean of three independent transfection experiments with bars indicating standard errors of the mean. Methylation status for each experiment was assessed using high-throughput bi-sulfite sequencing. Off-target TALE fusions bind to an unrelated sequence in the EGFP reporter gene. Note that the GFP control datapoints shown are the same for both graphs and are depicted twice for ease of comparison. (E) Demethylation activities of KL-2 TALE-TET1 CD (top) and KL-2 TALE-TET1 FL (bottom) fusion proteins in human K562 cells. Same as in (C) but using a TALE repeat array targeted to a second site in the KLF4 intron.
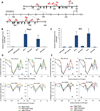
Figure 2. Targeted demethylation and activation of the human RHOXF2 gene by TALE-TET1 Fusion Proteins
(A) Schematic illustrating the human RHOXF2 locus with CpGs indicated with black arrows. Red arrows indicate the location and direction (5’ to 3’) of 11 TALE-TET1 fusion protein binding sites. Numbering indicates position on the DNA relative to the start site of transcription (right-angle arrow). (B) Expression levels of RHOXF2 mRNA (normalized to β-actin mRNA levels) in HeLa cells transfected with plasmids expressing RH-3 or RH-4 TALE-TET1 proteins, RH-3 or RH-4 TALE-TET1 proteins bearing mutations that inactivate TET1 catalytic function, or an off-target TALE-TET1 protein (targeted to KLF4) or a control GFP expression plasmid. Means of three independent samples each assayed three times by quantitative RT-PCR are shown with bars representing standard errors of the mean. Asterisks indicate samples with values significantly greater than those obtained with the off-target control as determined by a one-sided Welch’s t-test (n=3, p<0.05). (C) Expression levels of RHOXF2 mRNA (normalized to β-actin mRNA levels) in 293 cells transfected with plasmids expressing the indicated TALE-TET1, catalytically inactivated TALE-TET1 or off-target TALE-TET1 fusion protein or a control GFP expression plasmid. Data presented as in (B). (D) Demethylation activities of RH-3 and RH-4 TALE-TET1 fusion proteins in HeLa (top) and 293 (bottom) cells. Graphs show the fraction of CpGs methylated (y-axis) for different positions along the length of the RHOXF2 promoter (x-axis, numbered relative to the transcription start site) in cells transfected with plasmids expressing the RH-3, RH-4 (blue), or an off-target (to a site in the KLF4 gene) TALE-TET1 protein (orange) or GFP (green). Each data point represents the mean of three independent transfection experiments with bars indicating standard errors of the mean. Methylation status for each experiment was assessed using high-throughput bi-sulfite sequencing. Note that the GFP and off-target TALE-TET1 control data points shown are the same in both panels for each cell type and are depicted multiple times only for ease of comparison with experimental samples. Blue arrow indicates the location and direction (5’ to 3’) of the RH3 or RH4 TALE-TET1 binding sites. (E) Demethylation activities of catalytically inactive RH-3 and RH-4 TALE-TET1 fusion proteins in HeLa (top) and 293 (bottom) cells. Data represented as in (D) but here showing fraction of CpGs methylated in cells transfected with plasmids expressing RH-3 or RH-4 TALE-TET1 fusions bearing mutations that inactivate TET1 function (red). Note that RH-3, RH-4 and GFP data are the same as in (D) and are depicted again here for ease of comparison with catalytically inactive controls.
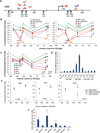
Figure 3. Targeted demethylation and activation of the human HBB gene by TALE-TET1 Fusion Proteins
(A) Schematic illustrating the human HBB locus with CpGs indicated with black arrows. Numbering indicates position on the DNA relative to the start site of transcription (right-angle arrow). Colored arrows indicate the location and direction (5’ to 3’) of 10 TALE-TET1 fusion protein binding sites. (B) Demethylation activities of HB-1, -2, -3, -4, -5, and -6 TALE-TET1 fusion proteins in K562 cells. Graphs show the fraction of CpGs methylated (y-axis) for different positions along the length of the HBB promoter (x-axis, numbered relative to the transcription start site) in K562 cells transfected with plasmids expressing one of the six HB TALE-TET1 fusion proteins (colored circles), an off-target TALE-TET1 fusion to a site in the KLF4 gene (black triangles), or GFP (green triangles). Each data point represents the mean of three independent transfection experiments with bars indicating standard errors of the mean. Methylation status for each experiment was assessed using high-throughput bisulfite sequencing. Note that the GFP and off-target TALE-TET1 control data points shown are the same in both panels and are depicted twice for ease of comparison with experimental samples. Colored arrows indicate the location and direction (5’ to 3’) of the various HB TALE-TET1 binding sites. (C) Demethylation activities of HB-7, -8, -9, and -10 TALE-TET1 fusion proteins in K562 cells. Data represented as in (B) (D) Expression levels of HBB mRNA (normalized to β-actin mRNA levels) in K562 cells transfected with indicated TALE-TET1 fusion protein expression plasmids or a control GFP expression plasmid. Means of three independent samples assayed by quantitative RT-PCR are shown with bars representing standard errors of the mean. Asterisks indicate samples with values significantly greater than those obtained with an off-target control TALE-TET1 fusion protein (targeted to a site in the human KLF4 gene) as determined by a one-sided Welch’s t-test (n=3, p<0.05). (E) Graphs showing the fraction of CpGs methylated (y-axis) for different positions along the length of the HBB promoter (x-axis, numbered relative to the transcription start site) in cells transfected with plasmids expressing the HB-4, HB-5 or HB-6 TALE-TET1 fusion, HB-4, HB-5 or HB-6 TALE-TET1 bearing mutations that inactivate TET1 catalytic function, or GFP. Each data point represents the mean of three independent transfection experiments with bars indicating standard errors of the mean. Methylation status for each experiment was assessed using high-throughput bi-sulfite sequencing. Note that the control GFP data shown is the same in all three panels and is depicted multiple times for ease of comparison with experimental samples. Blue arrow indicates the location and direction (5’ to 3’) of the TALE-TET1 fusion protein binding sites. (F) Expression levels of HBB mRNA (normalized to β-actin mRNA levels) in K562 cells transfected with plasmids expressing HB-4, HB-5 or HB-6 TALE-TET1 fusions, HB-4, HB-5 or HB-6 TALE-TET1 fusions bearing mutations that inactivate TET1 catalytic function, an off-target TALE-TET1 fusion targeted to an unrelated site in the human KLF4 gene, or GFP. Means of three independent samples assayed by quantitative RT-PCR are shown with bars representing standard errors of the mean. Asterisks indicate samples with values significantly greater than those obtained with the off-target control TALE-TET1 fusion protein as determined by a one-sided, two-sample equal variance t-test (n=3, p<0.05).
Comment in
- Epigenome editing.
Voigt P, Reinberg D. Voigt P, et al. Nat Biotechnol. 2013 Dec;31(12):1097-9. doi: 10.1038/nbt.2756. Nat Biotechnol. 2013. PMID: 24316647 No abstract available.
Similar articles
- Epigenome editing.
Voigt P, Reinberg D. Voigt P, et al. Nat Biotechnol. 2013 Dec;31(12):1097-9. doi: 10.1038/nbt.2756. Nat Biotechnol. 2013. PMID: 24316647 No abstract available. - Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions.
Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, Hatada I. Morita S, et al. Nat Biotechnol. 2016 Oct;34(10):1060-1065. doi: 10.1038/nbt.3658. Epub 2016 Aug 29. Nat Biotechnol. 2016. PMID: 27571369 - Editing DNA Methylation in the Mammalian Genome.
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Liu XS, et al. Cell. 2016 Sep 22;167(1):233-247.e17. doi: 10.1016/j.cell.2016.08.056. Cell. 2016. PMID: 27662091 Free PMC article. - Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain.
Gallego-Bartolomé J, Gardiner J, Liu W, Papikian A, Ghoshal B, Kuo HY, Zhao JM, Segal DJ, Jacobsen SE. Gallego-Bartolomé J, et al. Proc Natl Acad Sci U S A. 2018 Feb 27;115(9):E2125-E2134. doi: 10.1073/pnas.1716945115. Epub 2018 Feb 14. Proc Natl Acad Sci U S A. 2018. PMID: 29444862 Free PMC article. - The role of active DNA demethylation and Tet enzyme function in memory formation and cocaine action.
Alaghband Y, Bredy TW, Wood MA. Alaghband Y, et al. Neurosci Lett. 2016 Jun 20;625:40-6. doi: 10.1016/j.neulet.2016.01.023. Epub 2016 Jan 19. Neurosci Lett. 2016. PMID: 26806038 Free PMC article. Review.
Cited by
- Active enhancers: recent research advances and insights into disease.
Zhang J, Wang Q, Liu J, Duan Y, Liu Z, Zhang Z, Li C. Zhang J, et al. Biol Direct. 2024 Nov 12;19(1):112. doi: 10.1186/s13062-024-00559-x. Biol Direct. 2024. PMID: 39533395 Free PMC article. Review. - Epigenetic regulation of human FOXP3+ Tregs: from homeostasis maintenance to pathogen defense.
Yue Y, Ren Y, Lu C, Li P, Zhang G. Yue Y, et al. Front Immunol. 2024 Jul 31;15:1444533. doi: 10.3389/fimmu.2024.1444533. eCollection 2024. Front Immunol. 2024. PMID: 39144146 Free PMC article. Review. - Optimized Protocol for the Regulation of DNA Methylation and Gene Expression Using Modified dCas9-SunTag Platforms.
Morita S, Horii T, Hatada I. Morita S, et al. Methods Mol Biol. 2024;2842:155-165. doi: 10.1007/978-1-0716-4051-7_7. Methods Mol Biol. 2024. PMID: 39012594 - Designing Epigenome Editors: Considerations of Biochemical and Locus Specificities.
Yagci ZB, Kelkar GR, Johnson TJ, Sen D, Keung AJ. Yagci ZB, et al. Methods Mol Biol. 2024;2842:23-55. doi: 10.1007/978-1-0716-4051-7_2. Methods Mol Biol. 2024. PMID: 39012589 - Development of Locus-Directed Editing of the Epigenome from Basic Mechanistic Engineering to First Clinical Applications.
Rots MG, Jeltsch A. Rots MG, et al. Methods Mol Biol. 2024;2842:3-20. doi: 10.1007/978-1-0716-4051-7_1. Methods Mol Biol. 2024. PMID: 39012588
References
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. - PubMed
- Brena RM, Costello JF. Genome-epigenome interactions in cancer. Hum. Mol. Genet. 2007;16 Spec No 1:R96–R105. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 NS45776/NS/NINDS NIH HHS/United States
- P50 HG005550/HG/NHGRI NIH HHS/United States
- P30 NS045776/NS/NINDS NIH HHS/United States
- R01 CA169316/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HD045595/HD/NICHD NIH HHS/United States
- DP1 GM105378/DP/NCCDPHP CDC HHS/United States
- F32 GM105189/GM/NIGMS NIH HHS/United States
- DP1 GM105378/GM/NIGMS NIH HHS/United States
- R01 HD053808/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials