A conserved protein with AN1 zinc finger and ubiquitin-like domains modulates Cdc48 (p97) function in the ubiquitin-proteasome pathway - PubMed (original) (raw)
A conserved protein with AN1 zinc finger and ubiquitin-like domains modulates Cdc48 (p97) function in the ubiquitin-proteasome pathway
Bebiana Sá-Moura et al. J Biol Chem. 2013.
Abstract
Regulated protein degradation mediated by the ubiquitin-proteasome system (UPS) is critical to eukaryotic protein homeostasis. Often vital to degradation of protein substrates is their disassembly, unfolding, or extraction from membranes. These processes are catalyzed by the conserved AAA-ATPase Cdc48 (also known as p97). Here we characterize the Cuz1 protein (Cdc48-associated UBL/zinc finger protein-1), encoded by a previously uncharacterized arsenite-inducible gene in budding yeast. Cuz1, like its human ortholog ZFAND1, has both an AN1-type zinc finger (Zf_AN1) and a divergent ubiquitin-like domain (UBL). We show that Cuz1 modulates Cdc48 function in the UPS. The two proteins directly interact, and the Cuz1 UBL, but not Zf_AN1, is necessary for binding to the Cdc48 N-terminal domain. Cuz1 also associates, albeit more weakly, with the proteasome, and the UBL is dispensable for this interaction. Cuz1-proteasome interaction is strongly enhanced by exposure of cells to the environmental toxin arsenite, and in a proteasome mutant, loss of Cuz1 enhances arsenite sensitivity. Whereas loss of Cuz1 alone causes only minor UPS degradation defects, its combination with mutations in the Cdc48(Npl4-Ufd1) complex leads to much greater impairment. Cuz1 helps limit the accumulation of ubiquitin conjugates on both the proteasome and Cdc48, suggesting a possible role in the transfer of ubiquitylated substrates from Cdc48 to the proteasome or in their release from these complexes.
Keywords: ATPases; Cdc48; Cuz1; ER-associated Degradation; Proteasome; Ubiquitin; Yeast; Zinc Finger.
Figures
FIGURE 1.
Sequence features of yeast Cuz1/YNL155w and YOR052c. A, schematic depicting domain organization of Cuz1 and Yor052c. Proteasome-associated control elements (PACE) are found upstream of the corresponding genes. B, Cuz1 is evolutionarily conserved, containing an AN1-type zinc finger (Zf_AN1) domain in its N-terminal region. Aligned proteins were chosen based on the phylogenetic tree of Cuz1 orthologs from the Phylome database. Alignments were performed with ClustalOmega and edited with ESPript. The expected zinc-coordinating residues of the N-terminal Zf_AN1 domain are indicated with a filled circle (●). Except for Cuz1 and Q754N1, all other proteins possess a second Zf_AN1 domain; the putative metal-coordinating residues of this second domain are marked with a filled star (★). Proteins are from the following species: YEAST, S. cerevisiae; HUMAN, Homo sapiens; MOUSE, Mus musculus; DANIO, Danio rerio; ASPFU, Aspergillus fumigatus; PHANO, Phaeosphaeria nodorum SN15; NEUCR, Neurospora crassa; CHAGB, Chaetomium globosum; ASHGO, Ashbya gossypii; APLCA, Aplysia californica; STRPU, Strongylocentrotus purpuratus; TRICA, Tribolium castaneum; CIOIN, Ciona intestinalis. Secondary structure elements were added using ESPript based on the Protein Data Bank file of the obtained model for the ubiquitin-like domain. C, Cuz1 contains a C-terminal UBL. Protein fold and three-dimensional structure predictions were obtained using Phyre2 (43). Model includes fragment 161–265 in green and aligned to ubiquitin (Protein Data Bank 1UBQ) in blue using PyMol. Phyre2 output model was predicted based on human ubiquilin 3 (d1yqba1).
FIGURE 2.
Identification of Cuz1-binding proteins in vivo. A, proteins from a strain expressing FLAG-Cuz1 from the chromosomal CUZ1 locus, or an untagged control strain, were affinity purified on an anti-FLAG resin; 10% of the purified sample was resolved in a 10% SDS-PAGE gel followed by silver staining. B, the remaining sample from the immunopurification was subjected to LC-MS/MS analysis. Using spectral counts as a semi-quantitative index, the majority of proteins showed similar abundance in both samples (untagged versus FLAG-Cuz1). The table shows the proteins from the FLAG-Cuz1 purification that had a spectral count (SC) ratio ≥5-fold above the untagged control.
FIGURE 3.
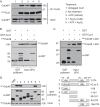
Cuz1 interacts directly with Cdc48. A, Cuz1 and Cdc48 associate in vivo, and ATP reduces their interaction. Cells expressed FLAG-tagged Cuz1 and Cdc48-V5 or, as a control, untagged Cuz1 and Cdc48-V5. Cultures were treated with 0.2 m
m
As2O3 for 2 h prior to lysis where indicated. ATP (2 m
m
) was added to the extracts where indicated. Following immunoprecipitation of FLAG-Cuz1, anti-V5 and anti-FLAG immunoblot analysis was performed. B, Cdc48 interaction with Cuz1 is direct. His6-Cdc48, GST, and GST-Cuz1 were expressed in and purified from E. coli. GST pulldowns were followed by anti-His tag and anti-GST immunoblotting. C, Cuz1 interacts with N-terminal domain of Cdc48. Binding of GST-Cuz1 to His6-Cdc48(1–220) was tested as described for the full-length construct in B. D, the Cuz1 Zf_AN1 domain is neither necessary nor sufficient for Cdc48 interaction, whereas the UBL domain is required. All proteins used in the GST pulldown analysis were purified from E. coli.
FIGURE 4.
Deletion of Cuz1 causes cellular protein degradation defects. A, pulse-chase analysis of UbVal-76-β-gal in the indicated yeast strains. Representative autoradiograph of a gel is shown at left. Arrowhead indicates a 90-kDa degradation product observed with UbVal-76-β-gal degradation in yeast. Bands above the primary UbVal-76-β-gal are polyubiquitylated species. The graph at right shows the mean degradation rates observed from three independent experiments. Error bars represent S.E. B, degradation of CPY*-HA was analyzed by cycloheximide chase/immunoblot analysis. CPY*-HA was detected by anti-HA immunoblotting. As a loading control, the membrane was subsequently probed with anti-PGK antibodies (bottom panels). C, growth assays reveal genetic interactions of _cuz1_Δ with mutations in Cdc48Npl4-Ufd1. 6-Fold serial dilutions of cultures were spotted onto plates (SD minimal medium or SD with 0.5 μg/ml of tunicamycin). The apparent growth advantage of cdc48-6 and _cdc48-6 cuz1_Δ in medium containing tunicamycin could in principle be due to a low constitutive induction of the ER unfolded-protein response in these cells. D, double mutant analysis of _cuz1_Δ with different UBX gene deletions. Myriocin was used at 0.2 μg/ml. 10-Fold serial dilutions of cultures were spotted onto the plates. Negative genetic interactions between _cuz1_Δ and _ubx1_Δ were also observed in a different strain background (not shown).
FIGURE 5.
Cuz1 associates with ubiquitin-protein conjugates in vivo. A, recombinant GST-Cuz1 was incubated with extracts from yeast overexpressing HA-tagged ubiquitin, and protein eluted from the glutathione resin was analyzed by anti-HA and anti-GST immunoblotting. B, the Cuz1 UBL domain is required for ubiquitin-conjugate interaction, but the Zf_AN1 domain is neither necessary nor sufficient. The indicated GST-tagged constructs were used in GST pulldown assays performed as in A. C, loss of Cuz1 enhances association of polyubiquitinated conjugates with Cdc48 in vivo. Analysis was done with cultures of CUZ1 or _cuz1_Δ cells expressing Cdc48-V5 from the endogenous CDC48 locus and expressing HA-tagged ubiquitin from a plasmid.
FIGURE 6.
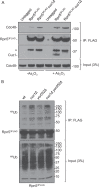
Genetic and physical interactions between Cuz1 and the proteasome. A, Cuz1 interacts in vivo with the 26 S proteasome. Proteasomes were affinity purified from extracts of yeast expressing either FLAG-tagged Pre1 (CP) or Rpn11 (RP) from the respective endogenous locus. The bound material was analyzed by anti-Cuz1 immunoblotting. B, interaction between Cuz1 and 26 S proteasomes in vitro. Recombinant GST-Cuz1 was incubated with yeast 26 S proteasomes affinity-purified from an Rpn5–3FLAG-expressing strain. C, the UBL domain is not required for Cuz1 interaction with proteasomes. Recombinant GST-Cuz1 constructs were mixed with purified proteasomal 19 S RP (purified from an Rpn11–3FLAG-expressing strain), and the proteins were subjected to GST pulldown analysis. D, deletion of CUZ1 exacerbates the growth defects of cim3-1 and cim5-1 proteasome mutants. Serial dilutions of cultures were done as described in the legend to Fig. 4_C_.
FIGURE 7.
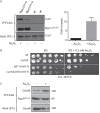
Cell exposure to arsenite enhances proteasome association with Cuz1. A, FLAG-tagged Rpn11 was immunoprecipitated and the amount of co-purifying Cuz1 was quantified using a Syngene G-box. A representative experiment is shown at the left. Cuz1 values were normalized to the levels of precipitated Rpn11–3FLAG. The quantification at the right is derived from three independent experiments and shows the normalized fold-increase of coprecipitated Cuz1; error bars denote S.D. B, in the presence of arsenite, deletion of CUZ1 worsens the growth defect of a temperature-sensitive cim5-1 mutant at high temperature. C, Cdc48 interaction with the proteasome is enhanced by arsenite. Proteasomes tagged with Rpn5–3FLAG were immunoprecipitated with anti-FLAG resin from extracts derived from cells grown in the presence of arsenite for 30 min. Co-purified Cdc48 levels was analyzed by anti-Cdc48 immunoblotting.
FIGURE 8.
Cuz1 affects the interaction of polyubiquitinated substrates with proteasomes. A, a slight increase in Cdc48 bound to the proteasome is observed in the absence of Cuz1, both in the presence and absence of arsenite. FLAG-tagged proteasomes were immunoprecipitated, and the amount of bound Cdc48 was analyzed by anti-Cdc48 immunoblotting. An unspecific band is indicated with an asterisk. B, an increase in polyubiquitylated proteins on the proteasome is observed when CUZ1 is deleted. After exposing cells for 2 h to 0.2 m
m
arsenite, cells were lysed and proteasomes were immunoprecipitated. Co-purified polyubiquitinated proteins were analyzed by anti-HA immunoblot analysis.
Similar articles
- Bidirectional substrate shuttling between the 26S proteasome and the Cdc48 ATPase promotes protein degradation.
Li H, Ji Z, Paulo JA, Gygi SP, Rapoport TA. Li H, et al. Mol Cell. 2024 Apr 4;84(7):1290-1303.e7. doi: 10.1016/j.molcel.2024.01.029. Epub 2024 Feb 23. Mol Cell. 2024. PMID: 38401542 - Cuz1/Ynl155w, a zinc-dependent ubiquitin-binding protein, protects cells from metalloid-induced proteotoxicity.
Hanna J, Waterman D, Isasa M, Elsasser S, Shi Y, Gygi S, Finley D. Hanna J, et al. J Biol Chem. 2014 Jan 17;289(3):1876-85. doi: 10.1074/jbc.M113.534032. Epub 2013 Dec 2. J Biol Chem. 2014. PMID: 24297164 Free PMC article. - In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome.
Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y. Tsuchiya H, et al. Mol Cell. 2017 May 18;66(4):488-502.e7. doi: 10.1016/j.molcel.2017.04.024. Mol Cell. 2017. PMID: 28525741 - Cdc48-Ufd1-Npl4: stuck in the middle with Ub.
Bays NW, Hampton RY. Bays NW, et al. Curr Biol. 2002 May 14;12(10):R366-71. doi: 10.1016/s0960-9822(02)00862-x. Curr Biol. 2002. PMID: 12015140 Review. - Cdc48/Shp1 participates in dissociation of protein complexes to regulate their activity.
Lauinger L, Flick K, Kaiser P. Lauinger L, et al. Curr Genet. 2021 Apr;67(2):263-265. doi: 10.1007/s00294-020-01136-1. Epub 2021 Jan 3. Curr Genet. 2021. PMID: 33388824 Review.
Cited by
- Tmc1 Is a Dynamically Regulated Effector of the Rpn4 Proteotoxic Stress Response.
Guerra-Moreno A, Hanna J. Guerra-Moreno A, et al. J Biol Chem. 2016 Jul 8;291(28):14788-95. doi: 10.1074/jbc.M116.726398. Epub 2016 May 12. J Biol Chem. 2016. PMID: 27226598 Free PMC article. - The Proteasome and Its Network: Engineering for Adaptability.
Finley D, Prado MA. Finley D, et al. Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a033985. doi: 10.1101/cshperspect.a033985. Cold Spring Harb Perspect Biol. 2020. PMID: 30833452 Free PMC article. Review. - A protein interaction map of the LSU processome.
McCann KL, Charette JM, Vincent NG, Baserga SJ. McCann KL, et al. Genes Dev. 2015 Apr 15;29(8):862-75. doi: 10.1101/gad.256370.114. Epub 2015 Apr 15. Genes Dev. 2015. PMID: 25877921 Free PMC article. - Role of Proteostasis Regulation in the Turnover of Stress Granules.
Hu R, Qian B, Li A, Fang Y. Hu R, et al. Int J Mol Sci. 2022 Nov 23;23(23):14565. doi: 10.3390/ijms232314565. Int J Mol Sci. 2022. PMID: 36498892 Free PMC article. Review. - Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Sun ZJ, Bhanu MK, Allan MG, Arthanari H, Wagner G, Hanna J. Sun ZJ, et al. PLoS One. 2016 Sep 23;11(9):e0163660. doi: 10.1371/journal.pone.0163660. eCollection 2016. PLoS One. 2016. PMID: 27662200 Free PMC article.
References
- Peters J. M., Franke W. W., Kleinschmidt J. A. (1994) Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 269, 7709–7718 - PubMed
- Schmidt M., Hanna J., Elsasser S., Finley D. (2005) Proteasome-associated proteins. Regulation of a proteolytic machine. Biol. Chem. 386, 725–737 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083050/GM/NIGMS NIH HHS/United States
- GM083050/GM/NIGMS NIH HHS/United States
- R37 GM046904/GM/NIGMS NIH HHS/United States
- GM046904/GM/NIGMS NIH HHS/United States
- R01 GM046904/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous