PINK1 is degraded through the N-end rule pathway - PubMed (original) (raw)
. 2013 Nov 1;9(11):1758-69.
doi: 10.4161/auto.24633. Epub 2013 Apr 17.
Affiliations
- PMID: 24121706
- PMCID: PMC4028335
- DOI: 10.4161/auto.24633
PINK1 is degraded through the N-end rule pathway
Koji Yamano et al. Autophagy. 2013.
Abstract
PINK1, a mitochondrial serine/threonine kinase, is the product of a gene mutated in an autosomal recessive form of Parkinson disease. PINK1 is constitutively degraded by an unknown mechanism and stabilized selectively on damaged mitochondria where it can recruit the E3 ligase PARK2/PARKIN to induce mitophagy. Here, we show that, under steady-state conditions, endogenous PINK1 is constitutively and rapidly degraded by E3 ubiquitin ligases UBR1, UBR2 and UBR4 through the N-end rule pathway. Following precursor import into mitochondria, PINK1 is cleaved in the transmembrane segment by a mitochondrial intramembrane protease PARL generating an N-terminal destabilizing amino acid and then retrotranslocates from mitochondria to the cytosol for N-end recognition and proteasomal degradation. Thus, sequential actions of mitochondrial import, PARL-processing, retrotranslocation and recognition by N-end rule E3 enzymes for the ubiquitin proteosomal degradation defines the rapid PINK1 turnover. PINK1 steady-state elimination by the N-end rule identifies a novel organelle to cytoplasm turnover pathway that yields a mechanism to flag damaged mitochondria for autophagic elimination.
Keywords: PARKIN; PARL; mitochondrial import; mitophagy; ubiquitin.
Figures
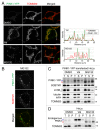
Figure 1. PARL-cleaved PINK1 can form cytosolic aggregates. (A and B) Microscopic analysis of HeLa cells transiently expressing PINK1-YFP treated with DMSO, valinomycin (Val) or MG132 for 3 h. Cells were immunostained with anti-TOMM20 (A) and with anti-SQSTM1 (B) antibodies. Line scan plots (taken from the images along the white arrows in (A) are shown in the right panel. Scale bars: 10 μm. (C and D) Immunoblotting of total cell lysate (T), supernatant (S) and pellet (P) fractions after solubilization with 2% Triton X-100 followed by centrifugation. Blue and red arrowheads represent the full-length and PARL-cleaved forms, respectively.
Figure 2. N-end rule pathway governs PINK1 degradation. (A) Scheme of PINK1-YFP variants. Bold downward arrows indicate the cleavage sites of PINK-YFP and Ub-PINK1-YFP by PARL and DUBs, respectively. (B and C) HeLa cells transiently expressing PINK1-YFP (B) or Ub-PINK1-YFP (C) variants were treated with DMSO or MG132 for 3 h. Total cell lysates were analyzed by immunoblotting. Tubulin, actin and TOMM20 were used as loading controls. (D) Proteasomal degradation efficiencies of cleaved forms of PINK1-YFP or Ub-PINK1-YFP variants prepared as in (B and C) were quantified. Data are the mean ± SD of three independent experiments. (E) Microscopy analysis of HeLa cells transiently expressing PINK1-YFP variants. Cells were immunostained with anti-TOMM20. Scale bars: 20 μm. (F) Quantification of localization of the PINK1-YFP mutated to stabilizing residues. Dual means that cells have a YFP signal both in mitochondria and in the cytosol. (G) Transfected cells as in (E) were fractionated. T, total fraction of post-nuclear supernatant; S, post-mitochondrial supernatant; M, mitochondrial fraction. Blue and red arrowheads denote full-length (or MPP-cleaved for R98F) and PARL-cleaved forms, respectively.
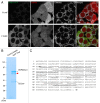
Figure 3. PARL cleaves PINK1F104M-YFP between A103 and M104. (A) HCT116 cells stably expressing wild-type (F104F) and the F104M mutant of PINK1-YFP-IRES-DsRed were immunostained with anti-TOMM20. Scale bars: 10 μm. (B) HCT116 cells stably expressing PINK1F104M-YFP were solubilized with 0.5% Triton X-100 and subjected to immunoprecipitation with GFP-Trap or control beads. Proteins were analyzed by SDS-PAGE followed by coomassie brilliant blue staining. Red arrowhead represents PARL-cleaved F104M mutant. Mass spectrometry identified HSP90AA1 and CDC37 as interacting partners of cleaved F104M mutant. (C) The protein band described by arrowhead in (B) was analyzed by LC/MS/MS, and the identified sequences of PINK1F104M moiety are underlined. Identifying the peptide corresponding to MGLGLGLIEEK indicates that PARL cleaves the PINK1F104M mutant between A103 and M104.
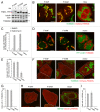
Figure 4. PINK1F104M mutant has the ability to recruit PARKIN to depolarized mitochondria followed by mitophagy. (A) Immunoblotting of HeLa cells expressing PINK1-YFP variants (F104F, F104M and 104Δ) treated with or without valinomycin (Val) for 3 h. Blue and red arrowheads represent the full-length and the cleaved forms of PINK1-YFP, respectively. (B and C) _pink1_−/− MEF cells were cotransfected with PINK1-YFP variants and mCherry-PARKIN. After 12 h of transfection, cells were treated with DMSO or valinomycin for 4 h. (B) Cells were then immunostained with anti-TOMM20. Scale bars: 10 μm. (C) Cells with mCherry-PARKIN on mitochondria were quantified. Error bars were calculated from three independent experiments. (D and E) pink1 KO HeLa cells stably expressing mCherry-PARKIN were cotransfected with YFP-LC3B and untagged either PINK1 WT(F104F) or PINK1(F104M) or pcDNA3.1(+) (vector). Cells were treated with valinomycin for 3 h, and immunostained with anti-TOMM20. (D) Scale bars: 10 μm. (E) Cells with YFP-LC3B on or near mitochondria were quantified. Error bars represent the results from three independent experiments. (F–I) pink1 KO HeLa cells stably expressing mCherry-PARKIN were transfected with WT PINK1-YFP (F104F), PINK1F104M-YFP or pEYFP-C1 (vector). Cells were treated with valinomycin for 12 h, and then immunostained with anti-TOMM20 or anti-HSPA9 antibodies. (F and H) YFP-positive cells are shown in white outlines. Scale bars: 10 μm. (G and I) Cells with complete degradation of TOMM20 or HSPA9 were quantified. Error bars represent the results from three independent experiments.
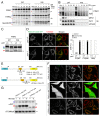
Figure 5. UBR proteins are responsible for PINK1 recognition. (A) Immunoblotting of total cell lysates from wild type (#1), _ubr1_−/−_ubr2_−/−_Luc_RNAi (#2) and _ubr1_−/−_ubr2_−/−_Ubr4_RNAi (#3) MEFs. (B) Total cell lysates from MEF cells cotransfected with PINK-YFP and mito-YFP at a 20:1 DNA ratio were analyzed by immunoblotting with anti-GFP antibody. In order to monitor any transfection efficiency discrepancies between the different cell lines, we used mito-YFP as an internal control, which was cotransfected with PINK1-YFP. The graph shows the relative amount of PARL-cleaved PINK1-YFP to mature mito-YFP, expressed as the mean ± SD of three independent experiments. Blue and red arrowheads denote full-length and PARL-cleaved PINK1-YFP. p and m denote precursor and mature forms of mito-YFP. (C) CCCP-treated MEF cells labeled with 35S-methionine/cysteine after PINK1-YFP transfection were subjected to CCCP-washout followed by chase for the indicated time periods. Immunoprecipitated PINK1-YFP was subjected to radioimaging. (D) Quantification of amounts of PARL-cleaved PINK1-YFP relative to full length PINK1-YFP at 0 time during the chase periods with the mean ± SD of three independent experiments. The amount of full-length PINK1-YFP at 0 time was set to 100%.
Figure 6. PINK1 precursor retrotranslocates from mitochondria to the cytosol after cleavage by PARL. (A) In vitro synthesized 35S-labeled proteins were incubated with mitochondria with or without valinomycin (Val). The mitochondrial pellet (pel) and the supernatant (sup) fractions were separated by centrifugation. 10% and 20% represent the amount of input of translation product. (B) PINK1 import samples were treated with PK and then the pel and sup were separated. The samples were subjected to radioimaging (low and high exposures) for PINK1 or immunoblotting (IB). (C) PINK1 import into _parl_−/− mitochondria. Blue, green and red arrowheads indicate the full-length, MPP-cleaved and PARL-cleaved PINK1, respectively. (D) Microscopic analysis of Parl+/+ and _parl_−/− MEF cells transiently expressing PINK1F104M-YFP. Cells were immunostained with anti-TOMM20. Scale bars: 10 μm. Right panel shows quantification of localization of PINK1-YFP wild-type (F104F), F104M and 104Δ mutants in Parl+/+ and _parl_−/− MEF cells. Dual means YFP signal both in mitochondria and in the cytosol. ND, not determined because of the low expression level. (E) Schematic representation of YFP-fusion proteins with N-terminal 155 residues of PINK1 or HTRA2. Arrows indicate the cleavage site by PARL for PINK1-YFP variants and by an IMS protease for HTRA2(1-155)-YFP. (F) Microscopic analysis of HeLa cells transiently expressing PINK1(1-155)-YFP, PINK1(1-155)F104M-YFP and HTRA2(1-155)-YFP. Cells were immunostained with anti-TOMM20. Scale bars: 10 μm. (G) The transfected cells as in (F) were treated with or without MG132 for 3 h. Total cell lysates were analyzed by immunoblotting with anti-GFP and anti-TOMM20 antibodies. Blue and red arrowheads represent precursor proteins and cleaved proteins, respectively.
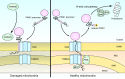
Figure 7. Schematic model of PINK1 trafficking to damaged or healthy mitochondria. Newly synthesized PINK1 precursor is accumulated on the outer membrane of damaged mitochondria, which can induce PARKIN recruitment from the cytosol to mitochondria. On the other hand, the N-terminal part of PINK1 precursor can be imported into the inner membrane of healthy mitochondria via TOM and TIMM23 translocator complexes. The N-terminal MTS and TM segment are cleaved by MPP and PARL, respectively. Subsequently, the cleaved PINK1 is released to the cytosol where the N-end rule specific E3 enzymes UBR1, UBR2 and UBR4 recognize the destabilizing N-terminal phenylalanine residue of cleaved PINK1 for proteasomal degradation.
Similar articles
- PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis.
Yan C, Gong L, Chen L, Xu M, Abou-Hamdan H, Tang M, Désaubry L, Song Z. Yan C, et al. Autophagy. 2020 Mar;16(3):419-434. doi: 10.1080/15548627.2019.1628520. Epub 2019 Jun 16. Autophagy. 2020. PMID: 31177901 Free PMC article. - Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy.
Meissner C, Lorenz H, Hehn B, Lemberg MK. Meissner C, et al. Autophagy. 2015;11(9):1484-98. doi: 10.1080/15548627.2015.1063763. Autophagy. 2015. PMID: 26101826 Free PMC article. - N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.
Eldeeb MA, Ragheb MA. Eldeeb MA, et al. Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review. - Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson's disease.
Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE. Shi G, et al. Hum Mol Genet. 2011 May 15;20(10):1966-74. doi: 10.1093/hmg/ddr077. Epub 2011 Feb 25. Hum Mol Genet. 2011. PMID: 21355049 - PINK1 import regulation at a crossroad of mitochondrial fate: the molecular mechanisms of PINK1 import.
Sekine S. Sekine S. J Biochem. 2020 Mar 1;167(3):217-224. doi: 10.1093/jb/mvz069. J Biochem. 2020. PMID: 31504668 Review.
Cited by
- Assessing Neuronal Mitophagy During Development in the Nematode Caenorhabditis elegans.
Ploumi C, Roussos A, Palikaras K. Ploumi C, et al. Methods Mol Biol. 2024;2845:55-66. doi: 10.1007/978-1-0716-4067-8_5. Methods Mol Biol. 2024. PMID: 39115657 - Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection.
Pellegrino MW, Haynes CM. Pellegrino MW, et al. BMC Biol. 2015 Apr 3;13:22. doi: 10.1186/s12915-015-0129-1. BMC Biol. 2015. PMID: 25857750 Free PMC article. Review. - Selective autophagy as a therapeutic target for neurological diseases.
Xu W, Ocak U, Gao L, Tu S, Lenahan CJ, Zhang J, Shao A. Xu W, et al. Cell Mol Life Sci. 2021 Feb;78(4):1369-1392. doi: 10.1007/s00018-020-03667-9. Epub 2020 Oct 16. Cell Mol Life Sci. 2021. PMID: 33067655 Free PMC article. Review. - The arginylation branch of the N-end rule pathway positively regulates cellular autophagic flux and clearance of proteotoxic proteins.
Jiang Y, Lee J, Lee JH, Lee JW, Kim JH, Choi WH, Yoo YD, Cha-Molstad H, Kim BY, Kwon YT, Noh SA, Kim KP, Lee MJ. Jiang Y, et al. Autophagy. 2016 Nov;12(11):2197-2212. doi: 10.1080/15548627.2016.1222991. Epub 2016 Aug 25. Autophagy. 2016. PMID: 27560450 Free PMC article. - The role of mitophagy in metabolic diseases and its exercise intervention.
Tang S, Geng Y, Lin Q. Tang S, et al. Front Physiol. 2024 Jan 29;15:1339128. doi: 10.3389/fphys.2024.1339128. eCollection 2024. Front Physiol. 2024. PMID: 38348222 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous