Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity - PubMed (original) (raw)
Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity
Kenneth A Matreyek et al. PLoS Pathog. 2013.
Abstract
Lentiviruses can infect non-dividing cells, and various cellular transport proteins provide crucial functions for lentiviral nuclear entry and integration. We previously showed that the viral capsid (CA) protein mediated the dependency on cellular nucleoporin (NUP) 153 during HIV-1 infection, and now demonstrate a direct interaction between the CA N-terminal domain and the phenylalanine-glycine (FG)-repeat enriched NUP153 C-terminal domain (NUP153(C)). NUP153(C) fused to the effector domains of the rhesus Trim5α restriction factor (Trim-NUP153(C)) potently restricted HIV-1, providing an intracellular readout for the NUP153(C)-CA interaction during retroviral infection. Primate lentiviruses and equine infectious anemia virus (EIAV) bound NUP153(C) under these conditions, results that correlated with direct binding between purified proteins in vitro. These binding phenotypes moreover correlated with the requirement for endogenous NUP153 protein during virus infection. Mutagenesis experiments concordantly identified NUP153(C) and CA residues important for binding and lentiviral infectivity. Different FG motifs within NUP153(C) mediated binding to HIV-1 versus EIAV capsids. HIV-1 CA binding mapped to residues that line the common alpha helix 3/4 hydrophobic pocket that also mediates binding to the small molecule PF-3450074 (PF74) inhibitor and cleavage and polyadenylation specific factor 6 (CPSF6) protein, with Asn57 (Asp58 in EIAV) playing a particularly important role. PF74 and CPSF6 accordingly each competed with NUP153(C) for binding to the HIV-1 CA pocket, and significantly higher concentrations of PF74 were needed to inhibit HIV-1 infection in the face of Trim-NUP153(C) expression or NUP153 knockdown. Correlation between CA mutant viral cell cycle and NUP153 dependencies moreover indicates that the NUP153(C)-CA interaction underlies the ability of HIV-1 to infect non-dividing cells. Our results highlight similar mechanisms of binding for disparate host factors to the same region of HIV-1 CA during viral ingress. We conclude that a subset of lentiviral CA proteins directly engage FG-motifs present on NUP153 to affect viral nuclear import.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
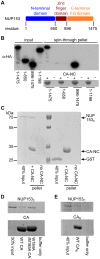
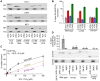
Figure 1. NUP153C directly binds the HIV-1 CA N-terminal domain.
(A) Schematic of NUP153 protein, with residue numbers of domain boundaries indicated. (B) Full length or truncated fragments of HA-tagged NUP153 extracted from 293T cells were tested for binding to HIV-1 CA-NC. Pelleted proteins were resolved by SDS-PAGE and visualized by western blotting with anti-HA antibody 3F10 (top), or by Coomassie stain (bottom). Input, 20% of binding reaction. CA-NC was included in the binding reactions as indicated. (C) Recombinant, tag-free NUP153C and GST purified from E. coli were similarly tested for binding to CA-NC; proteins were detected with Coomassie stain. (D) Recombinant NUP153C pulled down with full length his-tagged wild-type (WT) or W184A/M185A HIV-1 CA, and detected with Coomassie stain. (E) Recombinant NUP153C pulled down with his-tagged CAN, and detected with Coomassie stain. Each experiment was repeated at least 3 times, with a single representative result shown.
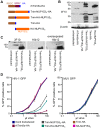
Figure 2. Restriction of HIV-1 infection by Trim5-NUP153C fusion proteins.
(A) Schematic of Trim-NUP153C fusion and control constructs. Color code: Trim5 RBCC, brown; rhTrim5α SPRY, auburn; HA-tag, blue; NUP153C, orange. (B) Western blot of HOS cells stably transduced with HA-tagged Trim-NUP153C fusion or control constructs, detected with antibody 3F10. (C) Western-blot detection of Trim-NUP153C fusion proteins with anti-HA monoclonal antibodies 3F10 and 16b12. Antibody 3F10 detects full-length Trim-NUP153C-HA whereas antibody 16b12 more faithfully detects full-length Trim-HA-NUP153C. (D) Infectivity of various doses of HIV-1 (left) or MLV (right) GFP reporter viruses on HOS cells stably expressing various Trim-based constructs. The results are an average of two experiments, with error bars denoting standard error. Asterisks in panels B and C mark bands that correspond to the expected mobilities of full length Trim-NUP153C constructs.
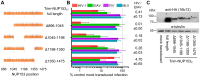
Figure 3. Diverse lentiviruses bind NUP153C.
(A) Transduction efficiencies of retroviral GFP reporter viruses in Trim-NUP153C expressing cells normalized to infection in mock transduced cells, which were set to 100%. Results are the geometric mean of 4 experiments, with error bars denoting 95% confidence intervals. (B) HA-NUP153C expressed in 293T cells was pulled down by the indicated his-tagged retroviral CAN proteins. Captured proteins resolved by SDS-PAGE were western blotted with antibody 3F10 alongside a standard curve of input protein. The results are an average of 5 experiments, with error bars denoting 95% confidence intervals. A representative western blot is shown. (C) SYPRO Ruby detection of retroviral CAN pull-down of purified NUP153C. The results are an average of two experiments, with error bars denoting standard error; one representative gel is shown. (D) (left) Retroviral infectivities in HOS cells knocked down for NUP153 expression as compared to cells treated with a non-targeting short interfering (si) RNA control . The results are the geometric mean of at least 4 experiments, with error bars denoting 95% confidence intervals. (right) Western blot detection of control or NUP153 knockdown HOS cells with antibody mab414, which also detects NUP358. (E) Scatter plot comparing relative retroviral infectivities under each condition.
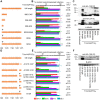
Figure 4. Different NUP153C sub-regions mediate Trim-NUP153C restriction of EIAV versus HIV-1 infection.
(A) To-scale schematic of NUP153C sequences encoded in various Trim-NUP153C constructs. Red lines represent boundaries of quarter-sized NUP153C sub-regions, while black lines denote the locations of FG motifs. (B) Infectivity of retroviral GFP reporter viruses on HOS cells stably expressing full-length or quarter-deleted Trim-NUP153C constructs, normalized to infection in mock transduced cells. Data represent the geometric mean of 5 experiments, with error bars denoting 95% confidence intervals. HIV-1 to EIAV ratios of infectivity are shown, with associated standard error. (C) Western blot of HOS cells stably transduced with Trim-NUP153C fusion constructs detected with antibody 16b12. Asterisks denote bands corresponding to the expected mobilities of full length or mutated Trim-NUP153C constructs.
Figure 5. The importance of FG motifs for Trim-NUP153C mediated inhibition of HIV-1 and EIAV infection.
To-scale schematics (A and D), normalized infection data (B and E), and western blotting (C and F) as described for Figure 4. Infection data are the geometric mean of at least 4 experiments, with error bars denoting 95% confidence intervals. Inverted grey triangle (panels A and D) denotes area of missense mutation.
Figure 6. FG motifs determine NUP153C binding to HIV-1 CAN.
(A) Pull-down of full-length or quarter deleted HA-NUP153C by HIV-1, EIAV, or MLV CAN proteins, detected with antibody 3F10. (B) Pull-down of WT NUP153C, FG-motif tetra-alanine mutant 1415A, or combinatorial 7×FG/A mutant by beads alone (none, grey), HIV-1 (red), MLV (blue), or EIAV (green) CAN proteins, as detected by western blot with antibody 3F10. Results are an average of at least 4 experiments, with error bars denoting standard error. (C) Purified NUP153C (black circles, solid line), NUP153CΔ1350–1475 (purple triangles, fine dotted line), and NUP153C7×FG/A (brown diamonds, coarse dotted line) proteins were incubated with various concentrations of HIV-1 CAN and a constant amount of Ni-NTA beads. Data points represent the mean and standard error of at least three experiments, fit with non-linear regression curves. The dissociation constant of NUP153C binding was calculated by averaging concentrations of half-maximal binding for 5 individual experiments, with associated standard error. (D) Sedimentation of WT NUP153C, FG-motif tetra-alanine mutant 1415A, combinatorial 7×FG/A mutant, or NUP153CΔ1350–1475 after incubation with buffer alone or assembled CA-NC. Results are an average of 6 experiments, with error bars denoting 95% confidence intervals. Representative western blotting results are shown.
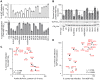
Figure 7. HIV-1 CA mutant-NUP153C binding and sensitivity to Trim-NUP153C restriction or NUP153 depletion.
(A) (top) Equal reverse transcriptase (RT) cpm of WT and HIV-1 mutant viruses plated on HOS cells, with resulting infectivities normalized to WT virus. (bottom) Percent infectivity of viruses in Trim-NUP153C expressing HOS cells, normalized to mock transduced control cells. Graphs show the mean of at least 5 experiments, with error bars denoting 95% confidence intervals. (B) Purified NUP153C pull-down by WT or the indicated mutant his-tagged HIV-1 CAN protein, with recovered proteins resolved by SDS-PAGE and detected by SYPRO Ruby stain. Results are an average of 5 experiments, with error bars denoting 95% confidence intervals. A representative staining result is shown. The dotted line highlights the level of NUP153C binding to WT CAN protein. (C) Scatter plot of NUP153C recovery in pull-down assays (panel B) compared to percent infectivity in Trim-NUP153C expressing cells (panel A, lower). Points are color-coded based on NUP153C binding phenotype: grey, not significantly different from WT; white, significantly decreased from WT; black, significantly increased from WT. (D) Scatter plot of normalized infectivity of CA mutant viruses in Trim-NUP153C expressing cells compared to the average infectivity of three experiments when endogenous NUP153 was knocked down. The comparison exhibited a significant Spearman rank correlation (P<0.0001). Points are color-coded as in panel C, except for CA mutants not tested for binding, which are denoted with “x” symbols.
Figure 8. NUP153C competes with molecules that bind the HIV-1 CAN hydrophobic pocket.
(A) X-ray crystal structure (pdb: 2×de) of compound PF74 (yellow) bound to HIV-1 CAN (green). Critical CAN side-chains (labeled) are shown as sticks, with oxygen and nitrogen atoms red and blue, respectively. Hydrogen bonds are shown as black dashes, with distances labeled. The phenylalanine moiety in PF74 is indicated by the black arrow. (B) PF74 competition of HA-NUP153C binding to WT or mutant his-tagged HIV-1 CAN. Recovered HA-NUP153C was detected with antibody 3F10 and quantitated alongside a standard curve of serially diluted HA-NUP153C-containing lysate. Baseline background signal observed with T54A/N57A CAN was subtracted, and values were normalized to that of the DMSO control (2% DMSO final concentration in each sample). Results are an average of at least 2 experiments, with error bars denoting standard error. Representative western blotting results are shown. (C) X-ray crystal structure (pdb: 4b4n) of a peptide from CPSF6 (backbone carbon atoms shown as grey sticks) bound to CAN (green) in the same orientation as in panel A. Side-chains and hydrogen bonds are represented as in panel A. The CPSF6 Phe321 side chain is indicated by the black arrow. (D) Binding of HA-tagged, full-length CPSF6 protein in 293T cell extract to HIV-1 CA-NC protein, and competition with purified NUP153C or mutants thereof. Results of 5 experiments were normalized to the level of CPSF6 binding observed in the absence of competing factors, with error bars denoting standard error. (E) Western blot of HOS cells stably expressing Trim-CPSF6358 or CPSF6358, detected with antibody 3F10. (F) CA mutant virus sensitivities to Trim-NUP153C (black) and Trim-CPSF6358 (red) restriction, as compared to cells transduced with an empty vector. Results are an average of at least 3 experiments, with error bars denoting 95% confidence intervals. (G) Scatter plot of CA mutant sensitivities to NUP153 knockdown compared with sensitivities to inhibition by CPSF6358.
Figure 9. PF74 counteracts HIV-1 similarly in the face of Trim-NUP153C restriction or NUP153 knockdown.
Mock transduced and Trim-NUP153C expressing (A) or non-targeting control and NUP153 knockdown (B) HOS cells were infected with equal RT-cpm of denoted viruses in the presence of various PF74 concentrations. Results are shown as infectivity normalized to vehicle only control cells (top), or vehicle only infection for each cell type (bottom) to calculate EC90 values. Dashed lines represent Trim-NUP153C or NUP153 knockdown results in panels A and B, respectively. Results are an average of at least 3 experiments, with error bars denoting standard error. Calculated EC90 values are displayed with standard error.
Figure 10. Mode of NUP153C binding to EIAV CA.
(A) Alignment of residues corresponding to HIV-1 CA Leu56 through Asn74 among various retroviruses. Residues that significantly affected HIV-1 CAN binding with NUP153C are highlighted in yellow. (B) Alignment of HIV-1 CAN (green, pdb: 3mge) and EIAV CAN (gray, pdb: 1eia), with side-chains surrounding the pocket shown as sticks. (C) Retroviral sensitivities to inhibition by PF74. Color codes: HIV-1, green; SIVmac, black; MLV, red; EIAV, orange; BIV, blue; FIV, purple. Results are an average of two experiments. (D) Retroviral sensitivities to inhibition by Trim-CPSF6358. Results are an average of 3 experiments. (E) Infectivity of RT-cpm matched EIAV GFP-reporter viruses carrying CA point mutations. Results are an average of 3 experiments. (F) Pull-down of purified NUP153C by EIAV point mutant CAN proteins. Results are an average of 2 experiments. (G) Sensitivity of EIAV CA point-mutant viruses to Trim-NUP153C. Results are an average of 4 experiments. Error bars in each panel denote standard error.
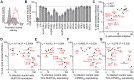
Figure 11. Association between NUP153 dependency and cell cycle independence.
(A) Propidium iodide staining of HOS cells untreated (grey) or treated for 24 h with 5 µM Etoposide phosphate (red line). (B) Infectivity of CA mutant viruses in HOS cells arrested with 5 µM Etoposide phosphate, normalized to infectivity in control HOS cells. Error bars denote standard error of 4 experiments. (C) Scatter plot comparison of CA mutant sensitivities in cell cycle arrested HOS cells in the absence or presence of 5 µM cyclosporine (CsA). Mutant viruses most sensitive to cell cycle arrest are indicated. (D to G) Scatter plots comparing sensitivities of mutant viruses to cell cycle arrest versus NUP153 knockdown (D), or restriction by Trim-NUP153C (E), CPSF6358 (F), or Trim-CPSF6358 (G). Spearman rank correlation coefficients and measures of significance are indicated. Data points for CA mutant viruses P38A, T54A, A92E, and G94D clustered with the WT virus within these panels, so their labels were omitted to aid legibility.
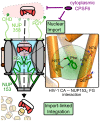
Figure 12. The NUP153-CA interaction during HIV-1 infection.
Partially uncoated HIV-1 cores dock at the NPC through engaging NUP358 (light green). Once docked, NUP153 (dark green) FG motifs bind CA through phenylalanine insertion into the hydrophobic pocket of the NTD, forming hydrogen bonds with CA residue Asn57, as well as adjacent polar side-chains (enlarged to the right). CA engagement with NUP153 is required for HIV-1 nuclear import, either directly during PIC translocation, or for completion of a prerequisite uncoating step. Perturbation of NUP153 engagement may affect multiple steps, such as intranuclear trafficking and integration site selection. Cytoplasmic CPSF6 may inhibit HIV-1 nuclear import by antagonizing NUP153 binding to CA.
Similar articles
- Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells.
Buffone C, Martinez-Lopez A, Fricke T, Opp S, Severgnini M, Cifola I, Petiti L, Frabetti S, Skorupka K, Zadrozny KK, Ganser-Pornillos BK, Pornillos O, Di Nunzio F, Diaz-Griffero F. Buffone C, et al. J Virol. 2018 Sep 12;92(19):e00648-18. doi: 10.1128/JVI.00648-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997211 Free PMC article. - Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly.
Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC. Price AJ, et al. PLoS Pathog. 2014 Oct 30;10(10):e1004459. doi: 10.1371/journal.ppat.1004459. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25356722 Free PMC article. - HIV-1 Resistance to the Capsid-Targeting Inhibitor PF74 Results in Altered Dependence on Host Factors Required for Virus Nuclear Entry.
Zhou J, Price AJ, Halambage UD, James LC, Aiken C. Zhou J, et al. J Virol. 2015 Sep;89(17):9068-79. doi: 10.1128/JVI.00340-15. Epub 2015 Jun 24. J Virol. 2015. PMID: 26109731 Free PMC article. - HIV Capsid and Integration Targeting.
Engelman AN. Engelman AN. Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review. - Capsid-host interactions for HIV-1 ingress.
Jang S, Engelman AN. Jang S, et al. Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004822. doi: 10.1128/mmbr.00048-22. Epub 2023 Sep 26. Microbiol Mol Biol Rev. 2023. PMID: 37750702 Free PMC article. Review.
Cited by
- A Branched SELEX Approach Identifies RNA Aptamers That Bind Distinct HIV-1 Capsid Structural Components.
Gruenke PR, Mayer MD, Aneja R, Schulze WJ, Song Z, Burke DH, Heng X, Lange MJ. Gruenke PR, et al. ACS Infect Dis. 2024 Aug 9;10(8):2637-2655. doi: 10.1021/acsinfecdis.3c00708. Epub 2024 Jul 17. ACS Infect Dis. 2024. PMID: 39016538 Free PMC article. - A Novel Phenotype Links HIV-1 Capsid Stability to cGAS-Mediated DNA Sensing.
Siddiqui MA, Saito A, Halambage UD, Ferhadian D, Fischer DK, Francis AC, Melikyan GB, Ambrose Z, Aiken C, Yamashita M. Siddiqui MA, et al. J Virol. 2019 Jul 30;93(16):e00706-19. doi: 10.1128/JVI.00706-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31167922 Free PMC article. - Regulation of Plant Immunity by Nuclear Membrane-Associated Mechanisms.
Fang Y, Gu Y. Fang Y, et al. Front Immunol. 2021 Dec 6;12:771065. doi: 10.3389/fimmu.2021.771065. eCollection 2021. Front Immunol. 2021. PMID: 34938291 Free PMC article. Review. - Quantitative monitoring of the cytoplasmic release of NCp7 proteins from individual HIV-1 viral cores during the early steps of infection.
Zgheib S, Lysova I, Réal E, Dukhno O, Vauchelles R, Pires M, Anton H, Mély Y. Zgheib S, et al. Sci Rep. 2019 Jan 30;9(1):945. doi: 10.1038/s41598-018-37150-0. Sci Rep. 2019. PMID: 30700731 Free PMC article. - Toward Structurally Novel and Metabolically Stable HIV-1 Capsid-Targeting Small Molecules.
Vernekar SKV, Sahani RL, Casey MC, Kankanala J, Wang L, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Vernekar SKV, et al. Viruses. 2020 Apr 16;12(4):452. doi: 10.3390/v12040452. Viruses. 2020. PMID: 32316297 Free PMC article.
References
- Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, et al. (1986) The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233: 215–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases