Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs - PubMed (original) (raw)
Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs
Dmytro Ustianenko et al. RNA. 2013 Dec.
Abstract
The mechanisms of gene expression regulation by miRNAs have been extensively studied. However, the regulation of miRNA function and decay has long remained enigmatic. Only recently, 3' uridylation via LIN28A-TUT4/7 has been recognized as an essential component controlling the biogenesis of let-7 miRNAs in stem cells. Although uridylation has been generally implicated in miRNA degradation, the nuclease responsible has remained unknown. Here, we identify the Perlman syndrome-associated protein DIS3L2 as an oligo(U)-binding and processing exoribonuclease that specifically targets uridylated pre-let-7 in vivo. This study establishes DIS3L2 as the missing component of the LIN28-TUT4/7-DIS3L2 pathway required for the repression of let-7 in pluripotent cells.
Keywords: DIS3L2; RNA degradation; RNA uridylation; let-7 miRNA.
Figures
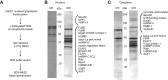
FIGURE 1.
Identification of Dis3l2 as an oligo(U)-binding nuclease in mouse embryonic stem cells (mESCs). (A) Schematic overview of the protocol. (B) Proteins identified in control and U30 RNA-bound nuclear extracts. (C) Proteins identified in control and U30 RNA-bound cytoplasmic extracts. Equal amounts of eluates from control RNA (Ctrl) and U30 RNA bait fractions were separated on 12% SDS-PAGE gels and silver stained. Proteins identified only in the U30 RNA sample are indicated on the right.
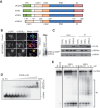
FIGURE 2.
DIS3L2 is a cytoplasmic, oligo(U)-binding, Mg++-dependent exoribonuclease that does not associate with exosomes. (A) Schematic representation of the domain organization of DIS3L2 homologs from Saccharomyces cerevisiae and Homo sapiens. The PIN domain is shown in green, the CSD1 and CSD2 RNA binding domains are in orange, the RNB ribonuclease domain is in blue, and the S1 domain is in pink. (B) Immunofluorescence staining of HEK293T and HeLa cells transfected with either DIS3L2 C-terminally fused to an EGFP (DIS3L2-EGFP) or an empty EGFP-expressing plasmid (EGFP). DAPI was used to visualize nuclei. The scale bar corresponds to 10 μm. The Western blot on the right shows the relative abundance of endogenous and EGFP-tagged DIS3L2 in transfected HeLa cells as detected with a DIS3L2-specific antibody. (C) DIS3L2 does not interact with core exosome components. FLAG-tagged DIS3, DIS3L, and DIS3L2 were immunoprecipitated from stable cell lines inducibly overexpressing the individual proteins. The composition of the IP samples was analyzed by Western blot with the indicated antibodies. (D) DIS3L2 binds U30 RNA with nanomolar affinity. Electromobility shift assay with recombinant DIS3L2 and 5′ end-32P-labeled U30 RNA. The migration patterns of free RNA and protein-RNA complexes are indicated. (E) The catalytic activity of DIS3L2 requires divalent metal cations. A degradation assay using recombinant DIS3L2 with U30 RNA as a substrate was performed in the presence of different divalent metal ions as indicated. (EDTA) Reaction mixture containing 5 mM EDTA; (np) control reaction without the addition of any protein.
FIGURE 3.
DIS3L2 targets 3′ end-uridylated precursors of let-7 miRNA. (A) Mutant DIS3L2 (D391N) specifically coprecipitates with extended forms of pre-let-7a miRNA. Northern blot analyses of RNAs coimmunoprecipitated with FLAG-tagged proteins as indicated on the upper side of the autoradiograph (IP-FLAG). The RNA input was total RNA isolated from whole cell lysates. The ethidium bromide (EtBr)-stained gels represent input loading controls. (Empty) Control RIP performed using cells that were not expressing any FLAG-tagged protein. The lower panel shows Western blot analysis of Myc-tagged LIN28A expression and the efficiency of FLAG tag-mediated immunoprecipitation of proteins used for RIP analyses. “Input” shows the whole cell lysates used for the IPs. The ectopic expression of Myc-tagged LIN28A was monitored with specific anti-Myc antibodies. DIS3L2 was detected with anti-FLAG antibodies. (B) Sequencing analysis of the 3′ termini of pre-let-7 RNAs coprecipitated with the D391N DIS3L2 mutant in HEK293T-Rex cells ectopically overexpressing LIN28A and pri-let-7a-1. The analysis suggests that the DIS3L2 mutant binds to uridylated pre-let-7a-1 and pre-let-7i because 31 out of 35 sequenced clones contained nontemplate oligo(U)-tails. The gray box schematically represents the body of pre-let-7 miRNAs. A graphical representation of the (U)-extension statistics is shown at the right. (C) Oligouridylation stimulates DIS3L2-mediated degradation of pre-let-7 miRNA. An in vitro degradation assay with FLAG-DIS3L2 purified from HEK293T-Rex cells and pre-let-7 RNA (represented by the schematic stem–loop) containing either no 3′ extension or 3′ oligo(U) and oligo(A) extensions of increasing lengths (indicated at the top) as substrates. D391N indicates a reaction with purified catalytically inactive DIS3L2. (D) DIS3L2 exhibits a preference for RNA substrates with oligo(U) extensions. A 5′ end-labeled pre-let-7-U8 RNA was incubated with purified DIS3L2 in the presence of a 10-fold excess of different unlabeled competitor RNAs (indicated at the top) for the time periods shown.
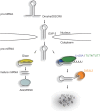
FIGURE 4.
A model for the regulation of let-7 miRNA biogenesis via the LIN28A-TUT4/7-DIS3L2 pathway. The primary transcripts (pri-let-7) are processed by the microprocessor complex in the nucleus to pre-let-7, which is exported to the cytoplasm. In the cytoplasm, pre-let-7 is either processed by Dicer to mature let-7 miRNA, or it is oligouridylated in the presence of LIN28A. The oligo(U) tails are then recognized by the DIS3L2 exonuclease, which initiates the degradation of RNA in the 3′ to 5′ direction.
Similar articles
- A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway.
Chang HM, Triboulet R, Thornton JE, Gregory RI. Chang HM, et al. Nature. 2013 May 9;497(7448):244-8. doi: 10.1038/nature12119. Epub 2013 Apr 17. Nature. 2013. PMID: 23594738 Free PMC article. - A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B.
Suzuki HI, Katsura A, Miyazono K. Suzuki HI, et al. Cancer Sci. 2015 Sep;106(9):1174-81. doi: 10.1111/cas.12721. Epub 2015 Jul 14. Cancer Sci. 2015. PMID: 26080928 Free PMC article. - Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway.
Faehnle CR, Walleshauser J, Joshua-Tor L. Faehnle CR, et al. Nature. 2014 Oct 9;514(7521):252-256. doi: 10.1038/nature13553. Epub 2014 Aug 3. Nature. 2014. PMID: 25119025 Free PMC article. - Regulation of RNA decay and cellular function by 3'-5' exoribonuclease DIS3L2.
Luan S, Luo J, Liu H, Li Z. Luan S, et al. RNA Biol. 2019 Feb;16(2):160-165. doi: 10.1080/15476286.2018.1564466. Epub 2019 Jan 13. RNA Biol. 2019. PMID: 30638126 Free PMC article. Review. - Biogenesis and regulation of the let-7 miRNAs and their functional implications.
Lee H, Han S, Kwon CS, Lee D. Lee H, et al. Protein Cell. 2016 Feb;7(2):100-13. doi: 10.1007/s13238-015-0212-y. Epub 2015 Sep 23. Protein Cell. 2016. PMID: 26399619 Free PMC article. Review.
Cited by
- To kill a microRNA: emerging concepts in target-directed microRNA degradation.
Buhagiar AF, Kleaveland B. Buhagiar AF, et al. Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review. - Dis3l2-Mediated Decay Is a Quality Control Pathway for Noncoding RNAs.
Pirouz M, Du P, Munafò M, Gregory RI. Pirouz M, et al. Cell Rep. 2016 Aug 16;16(7):1861-73. doi: 10.1016/j.celrep.2016.07.025. Epub 2016 Aug 4. Cell Rep. 2016. PMID: 27498873 Free PMC article. - Terminal Uridylyltransferases TUT4/7 Regulate microRNA and mRNA Homeostasis.
Zhang P, Frederick MI, Heinemann IU. Zhang P, et al. Cells. 2022 Nov 23;11(23):3742. doi: 10.3390/cells11233742. Cells. 2022. PMID: 36497000 Free PMC article. - Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection.
Yeh YJ, Tseng CP, Hsu SD, Huang HY, Lai MMC, Huang HD, Cheng JC. Yeh YJ, et al. J Virol. 2021 Jan 28;95(4):e01800-20. doi: 10.1128/JVI.01800-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33208444 Free PMC article. - H19 potentiates let-7 family expression through reducing PTBP1 binding to their precursors in cholestasis.
Zhang L, Yang Z, Huang W, Wu J. Zhang L, et al. Cell Death Dis. 2019 Feb 18;10(3):168. doi: 10.1038/s41419-019-1423-6. Cell Death Dis. 2019. PMID: 30778047 Free PMC article.
References
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284. - PubMed
- Bussing I, Slack FJ, Grosshans H. 2008. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 14: 400–409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials