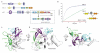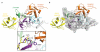Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP - PubMed (original) (raw)
. 2013 Nov 21;503(7476):422-426.
doi: 10.1038/nature12638. Epub 2013 Oct 20.
Affiliations
- PMID: 24141947
- PMCID: PMC3838313
- DOI: 10.1038/nature12638
Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP
Benjamin Stieglitz et al. Nature. 2013.
Abstract
Linear ubiquitin chains are important regulators of cellular signalling pathways that control innate immunity and inflammation through nuclear factor (NF)-κB activation and protection against tumour necrosis factor-α-induced apoptosis. They are synthesized by HOIP, which belongs to the RBR (RING-between-RING) family of E3 ligases and is the catalytic component of LUBAC (linear ubiquitin chain assembly complex), a multisubunit E3 ligase. RBR family members act as RING/HECT hybrids, employing RING1 to recognize ubiquitin-loaded E2 while a conserved cysteine in RING2 subsequently forms a thioester intermediate with the transferred or 'donor' ubiquitin. Here we report the crystal structure of the catalytic core of HOIP in its apo form and in complex with ubiquitin. The carboxy-terminal portion of HOIP adopts a novel fold that, together with a zinc-finger, forms a ubiquitin-binding platform that orients the acceptor ubiquitin and positions its α-amino group for nucleophilic attack on the E3∼ubiquitin thioester. The C-terminal tail of a second ubiquitin molecule is located in close proximity to the catalytic cysteine, providing a unique snapshot of the ubiquitin transfer complex containing both donor and acceptor ubiquitin. These interactions are required for activation of the NF-κB pathway in vivo, and they explain the determinants of linear ubiquitin chain specificity by LUBAC.
Figures
Fig. 1. Structure of the catalytic core of HOIP
a, Composition of LUBAC. Boxed: the crystallized catalytic core HOIPCBR-C, biochemical assays employed HOIPRBR-C. Below, schematic of new elements identified: CBR, ZF1, ZF2 and helical base. b, Single turnover assays showing that lack of RING1 reduces activity 6.8-fold (HOIPCBR-C, 0.050 min−1 versus HOIPRBR-C, 0.341 min−1). c-e, Ribbon representation of HOIPCBR-C with the helical base in grey, CBR in purple, ZF1 in cyan and ZF2 and β-hairpin in green, zinc ions as spheres, coordinating residues as ball and sticks and the catalytic cysteine in yellow. The structure represents HOIPCBR-C from the ubiquitin complex and includes regions disordered in the apo form.
Fig. 2. The HOIPCBR-C/ubiquitin transfer complex containing donor and acceptor ubiquitin
a, Ribbon representation of HOIPCBR-C in complex with the acceptor (orange) and donor (yellow) ubiquitin. HOIPCBR-C is shown in the same orientation as in Fig. 1e. The positions of C885, donor G76 and acceptor M1 are indicated. Insert, contacts made by HOIPCBR-C with donor and acceptor ubiquitin. The arrow shows the proximity between G76 of the donor and Sγ of C885. b, The HOIPCBR-C/ubiquitin complex with HOIPCBR-C shown in a surface representation to emphasize the spatial relationship between the 3 molecules.
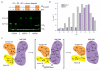
Fig. 3. Contacts between HOIPCBR-C and ubiquitin required for ubiquitin transfer
a, Close-up of the HOIPCBR-C/acceptor ubiquitin interface focusing on the helical base and ZF1. b, Details of the HOIPCBR-C/donor ubiquitin (yellow) interface. Positions of C885 and acceptor ubiquitin M1 are indicated. c, Steady-state ubiquitination assays. Mutants that target the acceptor interface are boxed in orange, those with donor in yellow. d, Single-turnover assays to determine the rate of tetraubiquitin formation. e, Luciferase assays showing that the NF-κB pathway is not efficiently activated by HOIP mutants H887A, R935A, D936A and D983A compared to WT. f, p65 translocations assay showing impaired p65 nuclear translocation upon expression of HOIP ligase-deficient mutants. Three independent experiments were performed using triplicate samples. Results were analyzed by ANOVA1 followed by Tukey post-tests. ** p<0.01; *** p<0.001 compared to wild-type HOIP.
Fig. 4. HOIP H887 acts as the general base to activate the nucleophile
a, HOIPRBR-C H887A and H889A (which shows WT activity) mutants can form a thioester (lanes 1 and 2 of each mutant), indicating that neither residue is involved in transthiolation from E2 to E3. However, H887A has lost the ability to transfer to a substrate to form diubiquitin. An orange square indicates the presence of acceptor ubiquitin. b, Single-turnover assays of the pH-dependence of tetraubiquitin formation showing that the H887A mutant regains activity at higher pH at 25°C. c, Proposed mechanism for ubiquitin transfer by HOIP.
Similar articles
- Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.
Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, Riedl SJ. Lechtenberg BC, et al. Nature. 2016 Jan 28;529(7587):546-50. doi: 10.1038/nature16511. Epub 2016 Jan 20. Nature. 2016. PMID: 26789245 Free PMC article. - The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension.
Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. Smit JJ, et al. EMBO J. 2012 Oct 3;31(19):3833-44. doi: 10.1038/emboj.2012.217. Epub 2012 Aug 3. EMBO J. 2012. PMID: 22863777 Free PMC article. - LUBAC synthesizes linear ubiquitin chains via a thioester intermediate.
Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. Stieglitz B, et al. EMBO Rep. 2012 Sep;13(9):840-6. doi: 10.1038/embor.2012.105. Epub 2012 Jul 13. EMBO Rep. 2012. PMID: 22791023 Free PMC article. - RBR E3 ubiquitin ligases: new structures, new insights, new questions.
Spratt DE, Walden H, Shaw GS. Spratt DE, et al. Biochem J. 2014 Mar 15;458(3):421-37. doi: 10.1042/BJ20140006. Biochem J. 2014. PMID: 24576094 Free PMC article. Review. - RBR E3-ligases at work.
Smit JJ, Sixma TK. Smit JJ, et al. EMBO Rep. 2014 Feb;15(2):142-54. doi: 10.1002/embr.201338166. Epub 2014 Jan 27. EMBO Rep. 2014. PMID: 24469331 Free PMC article. Review.
Cited by
- Linear ubiquitination signals in adaptive immune responses.
Ikeda F. Ikeda F. Immunol Rev. 2015 Jul;266(1):222-36. doi: 10.1111/imr.12300. Immunol Rev. 2015. PMID: 26085218 Free PMC article. Review. - Enzymatic Logic of Ubiquitin Chain Assembly.
Deol KK, Lorenz S, Strieter ER. Deol KK, et al. Front Physiol. 2019 Jul 5;10:835. doi: 10.3389/fphys.2019.00835. eCollection 2019. Front Physiol. 2019. PMID: 31333493 Free PMC article. Review. - Linkage reprogramming by tailor-made E3s reveals polyubiquitin chain requirements in DNA-damage bypass.
Wegmann S, Meister C, Renz C, Yakoub G, Wollscheid HP, Takahashi DT, Mikicic I, Beli P, Ulrich HD. Wegmann S, et al. Mol Cell. 2022 Apr 21;82(8):1589-1602.e5. doi: 10.1016/j.molcel.2022.02.016. Epub 2022 Mar 8. Mol Cell. 2022. PMID: 35263628 Free PMC article. - Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Xu X, Wang Y, Zhang Y, Wang Y, Yin Y, Peng C, Gong X, Li M, Zhang Y, Zhang M, Tang Y, Zhou X, Liu H, Pan L. Xu X, et al. Sci Adv. 2023 Oct 13;9(41):eadi4599. doi: 10.1126/sciadv.adi4599. Epub 2023 Oct 13. Sci Adv. 2023. PMID: 37831767 Free PMC article. - Selective localization of Mfn2 near PINK1 enables its preferential ubiquitination by Parkin on mitochondria.
Vranas M, Lu Y, Rasool S, Croteau N, Krett JD, Sauvé V, Gehring K, Fon EA, Durcan TM, Trempe JF. Vranas M, et al. Open Biol. 2022 Jan;12(1):210255. doi: 10.1098/rsob.210255. Epub 2022 Jan 19. Open Biol. 2022. PMID: 35042405 Free PMC article.
References
- Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi:ncb1821 [pii] 10.1038/ncb1821. - PubMed
- Tokunaga F, Iwai K. LUBAC, a novel ubiquitin ligase for linear ubiquitination, is crucial for inflammation and immune responses. Microbes Infect. 2012 doi:10.1016/j.micinf.2012.01.011. - PubMed
- Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi:10.1038/nature09816. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U117565398/MRC_/Medical Research Council/United Kingdom
- 250241/ERC_/European Research Council/International
- WT_/Wellcome Trust/United Kingdom
- MC_U117565398/MRC_/Medical Research Council/United Kingdom
- 094112/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases