Chromatin dynamics during cellular reprogramming - PubMed (original) (raw)
Review
Chromatin dynamics during cellular reprogramming
Effie Apostolou et al. Nature. 2013.
Abstract
Induced pluripotency is a powerful tool to derive patient-specific stem cells. In addition, it provides a unique assay to study the interplay between transcription factors and chromatin structure. Here, we review the latest insights into chromatin dynamics that are inherent to induced pluripotency. Moreover, we compare and contrast these events with other physiological and pathological processes that involve changes in chromatin and cell state, including germ cell maturation and tumorigenesis. We propose that an integrated view of these seemingly diverse processes could provide mechanistic insights into cell fate transitions in general and might lead to new approaches in regenerative medicine and cancer treatment.
Figures
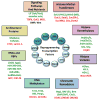
Figure 1. Dynamics of key epigenetic and transcriptional changes during direct reprogramming
Colored bars represent different cellular and transcriptional events (top panel) or epigenetic modifications (bottom panel) that change in dynamic patterns during iPSC formation. Examples of candidate enzymes that have been associated with these epigenetic marks in the context of direct reprogramming are given on the right. OKSM: Oct4, Klf4, Sox2, cMyc; 5mC: 5′-methyl-Cytosine; 5hmC: 5′-hydroxy-methyl-Cytosine; MET: Mesenchymal-to-epithelial-transition.
Figure 2. Interplay between reprogramming factors and molecules influencing chromatin state
Shown are different transcription factors that have been shown to trigger induced pluripotency, with the classical combination (Oct4, Sox2, Klf4, c-Myc) highlighted. Below each category are examples of molecules that have been shown to facilitate (green) or inhibit (red) reprogramming. BMPs (Bone Morphogenetic Factors) and Wnts have stage-dependent enhancing or suppressive roles during iPSC formation (black, see text for details). RBPs: RNA Binding Proteins.
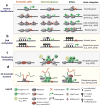
Figure 3. Levels of epigenetic gene regulation during induced pluripotency
a. Depicted are four categories of genes with associated histone modifications and their transcriptional response during reprogramming. Examples of genes within each category are shown to the right. b. Gain and loss of DNA methylation occurs late in reprogramming, while the acquisition of hydroxymethylation at pluripotency genes takes place at early-to-mid stages of iPSC formation. c. Oct4 (O), Klf4 (K) and Sox2 (S) function as “pioneer factors” that bind to high-density nucleosome regions, enabling chromatin remodeling and the recruitment of other factors including c-Myc (M). d. Changes in long-range chromatin interactions around the Nanog locus during iPSC formation. iPSC formation re-establishes a 3D chromatin network typical of ESCs, and this process depends on Mediator and Cohesin. Colored loops represent somatic-specific (orange), intermediate-specific (green) and pluripotency-specific (blue) _Nanog_-interacting loci.
Similar articles
- Epigenetics of cellular reprogramming.
Krishnakumar R, Blelloch RH. Krishnakumar R, et al. Curr Opin Genet Dev. 2013 Oct;23(5):548-55. doi: 10.1016/j.gde.2013.06.005. Epub 2013 Aug 12. Curr Opin Genet Dev. 2013. PMID: 23948105 Free PMC article. Review. - Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process?
Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Jullien J, et al. Nat Rev Mol Cell Biol. 2011 Jun 23;12(7):453-9. doi: 10.1038/nrm3140. Nat Rev Mol Cell Biol. 2011. PMID: 21697902 Free PMC article. Review. - Reprogramming chromatin.
Ehrensberger AH, Svejstrup JQ. Ehrensberger AH, et al. Crit Rev Biochem Mol Biol. 2012 Sep;47(5):464-82. doi: 10.3109/10409238.2012.697125. Epub 2012 Jul 3. Crit Rev Biochem Mol Biol. 2012. PMID: 22757592 Review. - Generation of human induced pluripotent stem cells using epigenetic regulators reveals a germ cell-like identity in partially reprogrammed colonies.
Goyal A, Chavez SL, Reijo Pera RA. Goyal A, et al. PLoS One. 2013 Dec 12;8(12):e82838. doi: 10.1371/journal.pone.0082838. eCollection 2013. PLoS One. 2013. PMID: 24349377 Free PMC article. - Epigenetics of reprogramming to induced pluripotency.
Papp B, Plath K. Papp B, et al. Cell. 2013 Mar 14;152(6):1324-43. doi: 10.1016/j.cell.2013.02.043. Cell. 2013. PMID: 23498940 Free PMC article. Review.
Cited by
- Dynamics of MBD2 deposition across methylated DNA regions during malignant transformation of human mammary epithelial cells.
Devailly G, Grandin M, Perriaud L, Mathot P, Delcros JG, Bidet Y, Morel AP, Bignon JY, Puisieux A, Mehlen P, Dante R. Devailly G, et al. Nucleic Acids Res. 2015 Jul 13;43(12):5838-54. doi: 10.1093/nar/gkv508. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007656 Free PMC article. - Inhibition of EZH2 primes the cardiac gene activation via removal of epigenetic repression during human direct cardiac reprogramming.
Tang Y, Zhao L, Yu X, Zhang J, Qian L, Jin J, Lu R, Zhou Y. Tang Y, et al. Stem Cell Res. 2021 May;53:102365. doi: 10.1016/j.scr.2021.102365. Epub 2021 Apr 27. Stem Cell Res. 2021. PMID: 34087994 Free PMC article. - Epigenetic regulation of stemness maintenance in the neurogenic niches.
Montalbán-Loro R, Domingo-Muelas A, Bizy A, Ferrón SR. Montalbán-Loro R, et al. World J Stem Cells. 2015 May 26;7(4):700-10. doi: 10.4252/wjsc.v7.i4.700. World J Stem Cells. 2015. PMID: 26029342 Free PMC article. Review. - Knockdown of Brm and Baf170, Components of Chromatin Remodeling Complex, Facilitates Reprogramming of Somatic Cells.
Jiang Z, Tang Y, Zhao X, Zhang M, Donovan DM, Tian XC. Jiang Z, et al. Stem Cells Dev. 2015 Oct 1;24(19):2328-36. doi: 10.1089/scd.2015.0069. Epub 2015 Jun 29. Stem Cells Dev. 2015. PMID: 26121422 Free PMC article. - The Third Intron of the Interferon Regulatory Factor-8 Is an Initiator of Repressed Chromatin Restricting Its Expression in Non-Immune Cells.
Khateb M, Fourier N, Barnea-Yizhar O, Ram S, Kovalev E, Azriel A, Rand U, Nakayama M, Hauser H, Gepstein L, Levi BZ. Khateb M, et al. PLoS One. 2016 Jun 3;11(6):e0156812. doi: 10.1371/journal.pone.0156812. eCollection 2016. PLoS One. 2016. PMID: 27257682 Free PMC article.
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources