Bacterial evolution of antibiotic hypersensitivity - PubMed (original) (raw)
doi: 10.1038/msb.2013.57.
Gajinder Pal Singh, Réka Spohn, István Nagy, Balázs Horváth, Mónika Hrtyan, Róbert Busa-Fekete, Balázs Bogos, Orsolya Méhi, Bálint Csörgő, György Pósfai, Gergely Fekete, Balázs Szappanos, Balázs Kégl, Balázs Papp, Csaba Pál
Affiliations
- PMID: 24169403
- PMCID: PMC3817406
- DOI: 10.1038/msb.2013.57
Bacterial evolution of antibiotic hypersensitivity
Viktória Lázár et al. Mol Syst Biol. 2013.
Abstract
The evolution of resistance to a single antibiotic is frequently accompanied by increased resistance to multiple other antimicrobial agents. In sharp contrast, very little is known about the frequency and mechanisms underlying collateral sensitivity. In this case, genetic adaptation under antibiotic stress yields enhanced sensitivity to other antibiotics. Using large-scale laboratory evolutionary experiments with Escherichia coli, we demonstrate that collateral sensitivity occurs frequently during the evolution of antibiotic resistance. Specifically, populations adapted to aminoglycosides have an especially low fitness in the presence of several other antibiotics. Whole-genome sequencing of laboratory-evolved strains revealed multiple mechanisms underlying aminoglycoside resistance, including a reduction in the proton-motive force (PMF) across the inner membrane. We propose that as a side effect, these mutations diminish the activity of PMF-dependent major efflux pumps (including the AcrAB transporter), leading to hypersensitivity to several other antibiotics. More generally, our work offers an insight into the mechanisms that drive the evolution of negative trade-offs under antibiotic selection.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
Networks of collateral-sensitivity interactions. Collateral-sensitivity interaction networks inferred from the adaptation to (A) low antibiotic concentrations and (B) increasing concentrations of antibiotics. Antibiotics are grouped according to their mode of action. An arrow from antibiotic A to antibiotic B indicates that adaptation to A increased the sensitivity to B. Aminoglycosides dominate the collateral-sensitivity network, with numerous links to other classes of antibiotics (red arrows). (C) Collateral-sensitivity antibiotic pairs show relatively low overlap in their chemogenomic profiles (_N_=120, Mann–Whitney _U_-test P<10−5). Chemogenomic distance was calculated as pairwise Jaccard distance between sets of genes that influence antibiotic susceptibility (Girgis et al, 2009). Error bars indicate 95% confidence intervals. (D) Collateral-sensitivity interaction degrees of antibiotic classes (i.e., average number of antibiotic classes against which a population evolves hypersensitivity if exposed to the antibiotic class shown on the vertical axis. Degrees are weighted by the number of antibiotics representing each class).
Figure 2
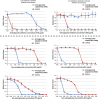
Dose–response curve of selected aminoglycoside-adapted lineages exhibiting collateral sensitivity. Error bars indicate 95% confidence intervals.
Figure 3
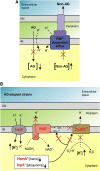
A putative mechanism underlying collateral sensitivity. (A) The theory. Altering the membrane potential across the inner bacterial membrane has two opposing effects: it reduces the uptake of many aminoglycoside-related antibiotics but simultaneously may lead to the reduced activity of PMF-dependent efflux pumps. For more details, see the main text. (B) Mutations supporting the theory. Whole-genome sequencing revealed that adaptation to aminoglycosides frequently proceeds through mutations that most likely diminish the generation of the PMF. Mutations are indicated by red, bolded protein names (TrkH, CyoB, HemA, IspA). The observed mutations in TrkH most likely increase the proton influx, whereas the mutations in CyoB and HemA (resulting in the inhibition of proton translocation and heme biosynthesis, respectively) interfere with the proper functioning of the cytochrome terminal oxidase complex. Furthermore, decreased IspA activity reduces the levels of membrane-bound quinones and therefore the level of oxidative phosphorylation. Altogether, these mutations likely reduce the PMF and thus aminoglycoside uptake. Simultaneously, the activity of the PMF-dependent efflux system is expected to decrease, resulting in greater sensitivity to antibiotics transported by these pumps. AG, aminoglycoside; OM, outer membrane; IM, inner membrane; NUO, NADH-Ubiquinone-oxidoreductase; PMF, proton-motive force.
Figure 4
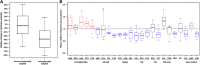
Membrane permeability (Hoechst dye) and membrane potential changes in evolved lines. (A) Membrane potential changes in antibiotic-adapted populations. Changes in the membrane potential were monitored using the carbocyanine dye DiOC2(3). The red/green fluorescence values for a representative set of aminoglycoside- and non-aminoglycoside-adapted populations were determined relative to the average of those of three wild-type controls. The membrane potential was significantly reduced in aminoglycoside-resistant populations (Wilcoxon rank-sum test _P_=0.002). Boxplots present the median and first and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data. The data are based on 10 and 22 measurements for aminoglycoside- and non-aminoglycoside-adapted populations, respectively. (B) Substantial differences in the accumulation of the fluorescent probe Hoechst 33342 across populations adapted to different classes of antibiotics (10 evolved populations each) relative to wild-type controls. Boxplots present the median and first and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (higher). The above figures are based on the results for lineages evolved in the presence of constant sublethal antibiotic concentrations. For further results, see Supplementary Figures S4 and S5.
Figure 5
Pleiotropic effects of a single mutation in trkH Individual antibiotic dose–response curves for growth inhibition were constructed for a trkH mutant strain. The red line denotes the trkH mutant strain and the blue line indicates the corresponding wild-type control. Error bars indicate the standard errors based on four technical replicates. A mutation in the trkH gene originally identified in a streptomycin-adapted population reduced the susceptibility to aminoglycosides but inhibited growth in the presence of several non-aminoglycoside antibiotic stresses. For more details on minimum inhibitory changes, see Supplementary Table S7. This mutation also (B) reduced the membrane potential (Wilcoxon rank-sum test _P_=0.02, based on four replicate measurements) and (C) the enhanced accumulation of Hoechst dye (Wilcoxon rank-sum test _P_=0.0005, based on eight replicate measurements). Control populations treated with a chemical inhibitor of the PMF (CCCP) showed similar patterns.
Figure 6
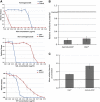
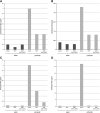
Link between the copy number of the major drug efflux system AcrAB and the extent of collateral-sensitivity interactions. The AcrAB efflux system confers resistance to a variety of drugs, but not to aminoglycosides. Two aminoglycoside-resistant strains (trkH* and TOB3) and a wild-type strain (control) were modified either by deleting the acrB gene (ΔacrB) or by harbouring a multicopy plasmid carrying the acrAB genes (pUCacrAB). Change of MIC in modified strains was measured using _E_-test stripes containing one of four antibiotics (A) Chloramphenicol, (B) Ciprofloxacin, (C) Doxycycline, (D) Trimethoprim, representing different classes of modes of action (Table I). The plasmid conferred a significant resistance to all four antibiotics in control strain, but resistance levels were substantially reduced when the same plasmid was associated with membrane potential affecting mutations in either trkH (trkH*) or both trkH and cyoB genes (TOB3).
References
- Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037–1050 - PubMed
- Baquero F (2001) Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat 4: 93–105 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical