C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci - PubMed (original) (raw)
C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci
Sarah Mizielinska et al. Acta Neuropathol. 2013 Dec.
Abstract
An expanded GGGGCC repeat in a non-coding region of the C9orf72 gene is a common cause of frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis. Non-coding repeat expansions may cause disease by reducing the expression level of the gene they reside in, by producing toxic aggregates of repeat RNA termed RNA foci, or by producing toxic proteins generated by repeat-associated non-ATG translation. We present the first definitive report of C9orf72 repeat sense and antisense RNA foci using a series of C9FTLD cases, and neurodegenerative disease and normal controls. A sensitive and specific fluorescence in situ hybridisation protocol was combined with protein immunostaining to show that both sense and antisense foci were frequent, specific to C9FTLD, and present in neurons of the frontal cortex, hippocampus and cerebellum. High-resolution imaging also allowed accurate analyses of foci number and subcellular localisation. RNA foci were most abundant in the frontal cortex, where 51 % of neurons contained foci. RNA foci also occurred in astrocytes, microglia and oligodendrocytes but to a lesser degree than in neurons. RNA foci were observed in both TDP-43- and p62-inclusion bearing neurons, but not at a greater frequency than expected by chance. RNA foci abundance in the frontal cortex showed a significant inverse correlation with age at onset of disease. These data establish that sense and antisense C9orf72 repeat RNA foci are a consistent and specific feature of C9FTLD, providing new insight into the pathogenesis of C9FTLD.
Figures
Fig. 1
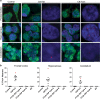
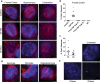
Sense RNA foci are a consistent and specific feature of C9FTLD. a RNA FISH for sense foci (red) was combined with immunostaining for neurons with NeuN (green) and nuclear DNA staining with DAPI (blue) in the frontal cortex, hippocampus and cerebellum. Representative images are shown from a neurologically normal control and heterozygous (C9 Het) and homozygous (C9 Hom) C9orf72 cases. Boxed regions are enlarged in the adjacent panel and only blue and red channels shown. Scale bar represents 5 μm in regular panels and 2 μm in enlarged panels. b Blind quantification for 10 normal controls, 8 C9FTLD cases, 4 FTLD-TDP type A cases, 3 FTLD-TDP type B cases (all without C9orf72 mutation) and 3 AD cases reveals sense RNA foci are specific to C9FTLD and present in all brain regions tested. Each dot represents an individual case with the homozygous C9FTLD case shown in red, and the average and SEM of heterozygous cases shown as long and short horizontal bars, respectively
Fig. 2
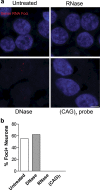
DNase and RNase treatments confirm specificity of RNA foci. a RNA FISH for sense foci (red) or CTG repeats [(CAG)7 probe, _bottom right panel_] was combined with nuclear DNA staining with DAPI (blue) in the hippocampus of the homozygous C9FTLD case. RNA foci were not detected after RNase treatment but were still present after DNase treatment, which was confirmed by blind quantification. b Loss of DAPI staining (bottom left panel) confirmed the DNase was effective. No signal was observed for the (CAG)7 probe, suggesting the sense RNA foci are not due to non-specific binding of the RNA probe. Scale bar represents 5 μm
Fig. 3
Localisation and frequency of sense RNA foci within neurons. RNA FISH for sense foci (red) was combined with immunostaining for neurons with NeuN (green) and nuclear DNA staining with DAPI (blue) in 8 C9FTLD cases. a The number of foci per neuron (in foci containing neurons) was quantified for each case in the frontal cortex (FC) hippocampus (Hc) and cerebellum (Cb). b Cumulative frequency distribution of number of foci per neuron in the 7 heterozygous C9FTLD cases (C9 Het, black bars) and the homozygous C9FTLD case (C9 Hom, red bars). c Neuron 1 is a frontal cortical neuron from the homozygous C9FTLD case containing 12 RNA foci (starred) including a cytoplasmic foci (white arrowhead). Neuron 2 is a frontal cortical neuron from a heterozygous C9FTLD case containing nuclear RNA foci (starred) which includes a RNA foci on the edge of the nucleus (arrowhead). Scale bar represents 2 μm. d Quantification of the percentage of neuronal RNA foci that are present on the edge of the nucleus and e within the cytoplasm. In a, d and e, each dot represents an individual C9FTLD case with the homozygous C9FTLD case shown in red, and the average and SEM of heterozygous cases shown as long and short horizontal bars, respectively. Significance was determined using one-way ANOVA and post hoc Bonferroni test (a and d) or paired t test (e): *p < 0.05, **p < 0.01, ***p < 0.001
Fig. 4
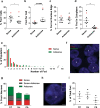
Sense RNA foci are present in the major glial subtypes. a RNA FISH for sense foci (red) was combined with immunostaining for astrocytes (GFAP), microglia (Iba1) or oligodendrocytes (CAII) (all in green), and DAPI (blue) in the frontal cortex of 8 C9FTLD cases. Scale bar represents 2 μm. The percentage of each cell type containing foci and the number of foci per cell are quantified in (b) and (c), respectively. d Graphical representation of the localisation of sense RNA foci in each cell type. In b, c, each dot represents an individual C9FTLD case with the homozygous C9FTLD case shown in red, and the average and SEM of heterozygous cases shown as long and short horizontal bars, respectively. Significance was determined using the one-way ANOVA and post hoc Bonferroni test (b, c): *p < 0.05, ***p < 0.001, ****p < 0.0001
Fig. 5
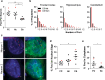
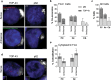
Antisense RNA foci are a consistent and specific feature of C9FTLD. RNA FISH for antisense foci (green) was combined with immunostaining for neurons with NeuN (red) and nuclear DNA staining with DAPI (blue). a Representative images from the frontal cortex, hippocampus and cerebellum from a neurologically normal control and heterozygous (C9 Het) and homozygous (C9 Hom) C9FTLD cases. Blind quantification of antisense RNA foci in frontal cortical neurons in C9FTLD cases and controls (b) and granule cell neurons of the hippocampus and cerebellum in C9FTLD cases only (c). d Representative images from a heterozygous C9FTLD case show antisense RNA foci are present in astrocytes, microglia, and oligodendrocytes. e RNA FISH for antisense foci was combined with RNase or DNase treatment, confirming the antisense probe detects RNA and not DNA. Scale bar represents 2 μm in all panels. In b, c, each dot represents an individual case with the homozygous C9FTLD case shown in red, and the average and SEM of heterozygous cases shown as long and short horizontal bars, respectively
Fig. 6
Comparison of sense and antisense RNA foci. a–e RNA FISH for either sense foci or antisense foci was combined with NeuN and DAPI staining in the frontal cortex of 8 C9FTLD cases. The percentage of neurons containing each foci type (a), the number of foci per cell (b), and their locations (c) and (d) were compared. e Frequency distribution of number of sense foci (red bars) and antisense foci (green bars) per neuron in the 7 heterozygous C9FTLD cases. The maximum number of sense RNA foci in a single neuron was 10 compared with 60 for antisense foci. f–i Sequential labelling of both sense and antisense foci was combined with NeuN and DAPI staining in a subset of C9FTLD cases in the frontal cortex (FC), hippocampus (Hc) and cerebellum (Cb). f Representative image from the hippocampus of the homozygous C9FTLD case showing neurons containing exclusively antisense foci (neuron 1), exclusively sense foci (neuron 2) or both antisense and sense foci (neuron 3). Scale bar represents 5 μm. g Quantification of the percentage of neurons containing sense foci, antisense foci, or both. h A neuron from the hippocampus of a heterozygous C9FTLD case showing co-localisation of sense and antisense foci (arrowed). Insets show enlarged region containing arrowed foci with individual channels for foci type and DAPI. Scale bar represents 2 μm. i Quantification of sense and antisense co-localisation from 3 heterozygous C9FTLD cases in all 3 brain regions. In a–d and i, each dot represents an individual C9FTLD case with the homozygous C9FTLD case shown in red, and the average and SEM of heterozygous cases shown as long and short horizontal bars, respectively. Significance was determined using a paired t test: *p < 0.05, **p < 0.01
Fig. 7
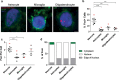
Comparison of RNA foci, p62 and TDP-43 inclusions in C9FTLD patient brain. RNA FISH for either sense or antisense foci was combined with immunostaining for p62 or phospho TDP-43 in the frontal cortex (FC), hippocampus (Hc) and cerebellum (Cb) of 3 heterozygous C9FTLD cases. a Representative images from the frontal cortex show that both sense and antisense foci occur in the same neurons as either p62-positive (TDP-43 negative) or TDP-43 inclusion pathology (both shown in white). b Quantification of co-occurrence of sense or antisense foci with p62 or TDP-43 pathology within the same cell in the frontal cortex, hippocampus and cerebellum shows no obvious associations between pathologies when compared to relative frequencies of each inclusion pathology in the total cell population in respective brain regions (c). d Representative images show that cytoplasmic sense foci (arrowed) can occasionally be found to co-localise within both p62 and TDP-43 inclusions in the hippocampus. Insets show enlarged region containing arrowed foci for each image without channel for p62 or TDP-43 to show clearly that foci are outside the nucleus. e Quantification of the co-localisation of cytoplasmic sense or antisense foci with either p62 or TDP-43 inclusions in the frontal cortex and hippocampus. Scale bars represent 2 μm. Data in graphs is shown as the average and SEM
Fig. 8
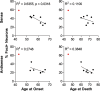
Clinical phenotypes correlate with the percentage of foci containing neurons in the frontal cortex of C9FTLD patient brain. Age at onset of disease or age at death was plotted against percentage of sense or antisense foci containing neurons in the frontal cortex for each individual case. Linear regressions were performed on data from heterozygous C9FTLD cases, with R 2 and any significant p value shown as a measure of goodness-of-fit and significance of the linear trend. Each dot represents an individual C9FTLD case with the homozygous C9FTLD case shown in red, even though it is not included in the linear regression analysis
Comment in
- Making sense of the antisense transcripts in C9FTD/ALS.
Todd PK. Todd PK. Acta Neuropathol. 2013 Dec;126(6):785-7. doi: 10.1007/s00401-013-1201-y. Acta Neuropathol. 2013. PMID: 24178412 Free PMC article. No abstract available.
Similar articles
- Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration.
Mizielinska S, Ridler CE, Balendra R, Thoeng A, Woodling NS, Grässer FA, Plagnol V, Lashley T, Partridge L, Isaacs AM. Mizielinska S, et al. Acta Neuropathol Commun. 2017 Apr 18;5(1):29. doi: 10.1186/s40478-017-0432-x. Acta Neuropathol Commun. 2017. PMID: 28420437 Free PMC article. - Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy.
Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, Pickering-Brown S, Kirby J, Hautbergue GM, Shaw PJ. Cooper-Knock J, et al. Acta Neuropathol. 2015 Jul;130(1):63-75. doi: 10.1007/s00401-015-1429-9. Epub 2015 May 6. Acta Neuropathol. 2015. PMID: 25943887 Free PMC article. - Drosha inclusions are new components of dipeptide-repeat protein aggregates in FTLD-TDP and ALS C9orf72 expansion cases.
Porta S, Kwong LK, Trojanowski JQ, Lee VM. Porta S, et al. J Neuropathol Exp Neurol. 2015 Apr;74(4):380-7. doi: 10.1097/NEN.0000000000000182. J Neuropathol Exp Neurol. 2015. PMID: 25756586 Free PMC article. - Mechanisms of toxicity in C9FTLD/ALS.
Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Gendron TF, et al. Acta Neuropathol. 2014 Mar;127(3):359-76. doi: 10.1007/s00401-013-1237-z. Epub 2014 Jan 7. Acta Neuropathol. 2014. PMID: 24394885 Free PMC article. Review. - The neuropathology associated with repeat expansions in the C9ORF72 gene.
Mackenzie IR, Frick P, Neumann M. Mackenzie IR, et al. Acta Neuropathol. 2014 Mar;127(3):347-57. doi: 10.1007/s00401-013-1232-4. Epub 2013 Dec 20. Acta Neuropathol. 2014. PMID: 24356984 Review.
Cited by
- Identification of selective and non-selective C9ORF72 targeting in vivo active siRNAs.
Gilbert JW, Kennedy Z, Godinho BMDC, Summers A, Weiss A, Echeverria D, Bramato B, McHugh N, Cooper D, Yamada K, Hassler M, Tran H, Gao FB, Brown RH Jr, Khvorova A. Gilbert JW, et al. Mol Ther Nucleic Acids. 2024 Jul 31;35(3):102291. doi: 10.1016/j.omtn.2024.102291. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39233852 Free PMC article. - Elucidating the Role of Cerebellar Synaptic Dysfunction in C9orf72-ALS/FTD - a Systematic Review and Meta-Analysis.
Kaliszewska A, Allison J, Col TT, Shaw C, Arias N. Kaliszewska A, et al. Cerebellum. 2022 Aug;21(4):681-714. doi: 10.1007/s12311-021-01320-0. Epub 2021 Sep 7. Cerebellum. 2022. PMID: 34491551 Free PMC article. Review. - Frontotemporal lobar degeneration: Pathogenesis, pathology and pathways to phenotype.
Mann DMA, Snowden JS. Mann DMA, et al. Brain Pathol. 2017 Nov;27(6):723-736. doi: 10.1111/bpa.12486. Epub 2017 Mar 2. Brain Pathol. 2017. PMID: 28100023 Free PMC article. Review. - C9orf72 poly GA RAN-translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity.
Lee YB, Baskaran P, Gomez-Deza J, Chen HJ, Nishimura AL, Smith BN, Troakes C, Adachi Y, Stepto A, Petrucelli L, Gallo JM, Hirth F, Rogelj B, Guthrie S, Shaw CE. Lee YB, et al. Hum Mol Genet. 2017 Dec 15;26(24):4765-4777. doi: 10.1093/hmg/ddx350. Hum Mol Genet. 2017. PMID: 28973350 Free PMC article. - C9ORF72: What It Is, What It Does, and Why It Matters.
Smeyers J, Banchi EG, Latouche M. Smeyers J, et al. Front Cell Neurosci. 2021 May 5;15:661447. doi: 10.3389/fncel.2021.661447. eCollection 2021. Front Cell Neurosci. 2021. PMID: 34025358 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- G1000287/MRC_/Medical Research Council/United Kingdom
- G1100695/MRC_/Medical Research Council/United Kingdom
- ISAACS/APR13/818-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/J004022/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources