A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control - PubMed (original) (raw)
doi: 10.7554/eLife.01123.
Jonathan M Carlson, Chanson J Brumme, Paul J McLaren, Zabrina L Brumme, Mina John, David W Haas, Javier Martinez-Picado, Judith Dalmau, Cecilio López-Galíndez, Concepción Casado, Andri Rauch, Huldrych F Günthard, Enos Bernasconi, Pietro Vernazza, Thomas Klimkait, Sabine Yerly, Stephen J O'Brien, Jennifer Listgarten, Nico Pfeifer, Christoph Lippert, Nicolo Fusi, Zoltán Kutalik, Todd M Allen, Viktor Müller, P Richard Harrigan, David Heckerman, Amalio Telenti, Jacques Fellay
Affiliations
- PMID: 24171102
- PMCID: PMC3807812
- DOI: 10.7554/eLife.01123
A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control
István Bartha et al. Elife. 2013.
Abstract
HIV-1 sequence diversity is affected by selection pressures arising from host genomic factors. Using paired human and viral data from 1071 individuals, we ran >3000 genome-wide scans, testing for associations between host DNA polymorphisms, HIV-1 sequence variation and plasma viral load (VL), while considering human and viral population structure. We observed significant human SNP associations to a total of 48 HIV-1 amino acid variants (p<2.4 × 10(-12)). All associated SNPs mapped to the HLA class I region. Clinical relevance of host and pathogen variation was assessed using VL results. We identified two critical advantages to the use of viral variation for identifying host factors: (1) association signals are much stronger for HIV-1 sequence variants than VL, reflecting the 'intermediate phenotype' nature of viral variation; (2) association testing can be run without any clinical data. The proposed genome-to-genome approach highlights sites of genomic conflict and is a strategy generally applicable to studies of host-pathogen interaction. DOI:http://dx.doi.org/10.7554/eLife.01123.001\.
Keywords: HIV; Human; human genomics; viral mutations.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
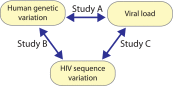
Figure 1.. A triangle of association testing.
The following association analyses were performed: [Study A] human SNPs vs plasma viral load (1 GWAS); [Study B] human SNPs vs variable HIV-1 amino acids (3007 GWAS); and [Study C] variable HIV-1 amino acids vs plasma viral load (1 proteome-wide association study). DOI:
http://dx.doi.org/10.7554/eLife.01123.003
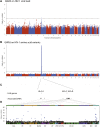
Figure 2.. Results of the genome-wide association analyses.
(A) Associations between human SNPs and HIV-1 plasma viral load. The dotted line shows the Bonferroni-corrected significance threshold (p-value < 7.25 × 10−9). (B) Associations between human SNPs and HIV-1 amino acid variants, with 3007 GWAS collapsed in a single Manhattan plot. The dotted line shows the Bonferroni-corrected significance threshold (p-value < 2.4 × 10−12). (C) Schematic representation of the HLA class I genes and of the SNPs associated with HIV-1 amino acid variants in the region. (D) Same association results as in panel B, projected on the HIV-1 proteome. Only the strongest association is shown for each amino acid. Significant associations are indicated by a blue dot. The gp120 part of the HIV-1 proteome was not tested. The colored bar below the plot area shows the positions of the optimally defined CD8+ T cell epitopes. An interactive version of this figure can be found at
(which is also available to download from Zenodo,
http://dx.doi.org/10.5281/zenodo.7138
). DOI:
http://dx.doi.org/10.7554/eLife.01123.004
Figure 3.. Association of HIV-1 amino acid variants with plasma viral load.
(A) Changes in VL (slope coefficients from the univariate regression model and standard error, log10 copies/ml) for the 48 HIV-1 amino acids that are associated with host SNPs in the genome-to-genome analysis. (B) rs2395029, a marker of HLA-B*57:01 is associated with a 0.38 log10 copies/ml lower VL (black bar) in comparison to the population mean. Gray bars represent changes in VL for amino acid variants associated with rs2395029 (p<0.001). In case of multiallelic positions, the change in VL is shown for all minor amino acids combined vs the major amino acid (e.g., GAG147 not I). DOI:
http://dx.doi.org/10.7554/eLife.01123.006
Similar articles
- Association Between Single-Nucleotide Polymorphisms in HLA Alleles and Human Immunodeficiency Virus Type 1 Viral Load in Demographically Diverse, Antiretroviral Therapy-Naive Participants From the Strategic Timing of AntiRetroviral Treatment Trial.
Ekenberg C, Tang MH, Zucco AG, Murray DD, MacPherson CR, Hu X, Sherman BT, Losso MH, Wood R, Paredes R, Molina JM, Helleberg M, Jina N, Kityo CM, Florence E, Polizzotto MN, Neaton JD, Lane HC, Lundgren JD. Ekenberg C, et al. J Infect Dis. 2019 Sep 13;220(8):1325-1334. doi: 10.1093/infdis/jiz294. J Infect Dis. 2019. PMID: 31219150 Free PMC article. Clinical Trial. - Human Immunotypes Impose Selection on Viral Genotypes Through Viral Epitope Specificity.
Gabrielaite M, Bennedbæk M, Zucco AG, Ekenberg C, Murray DD, Kan VL, Touloumi G, Vandekerckhove L, Turner D, Neaton J, Lane HC, Safo S, Arenas-Pinto A, Polizzotto MN, Günthard HF, Lundgren JD, Marvig RL. Gabrielaite M, et al. J Infect Dis. 2021 Dec 15;224(12):2053-2063. doi: 10.1093/infdis/jiab253. J Infect Dis. 2021. PMID: 33974707 Free PMC article. - Using viral diversity to identify HIV-1 variants under HLA-dependent selection in a systematic viral genome-wide screen.
Neuner-Jehle N, Zeeb M, Thorball CW, Fellay J, Metzner KJ, Frischknecht P, Neumann K, Leeman C, Rauch A, Stöckle M, Huber M, Perreau M, Bernasconi E, Notter J, Hoffmann M, Leuzinger K, Günthard HF, Pasin C, Kouyos RD; Swiss HIV Cohort Study (SHCS). Neuner-Jehle N, et al. PLoS Pathog. 2024 Aug 8;20(8):e1012385. doi: 10.1371/journal.ppat.1012385. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39116192 Free PMC article. - The impact of host genetic variation on infection with HIV-1.
McLaren PJ, Carrington M. McLaren PJ, et al. Nat Immunol. 2015 Jun;16(6):577-83. doi: 10.1038/ni.3147. Nat Immunol. 2015. PMID: 25988890 Free PMC article. Review. - HIV-1 dynamics: a reappraisal of host and viral factors, as well as methodological issues.
Prentice HA, Tang J. Prentice HA, et al. Viruses. 2012 Oct 10;4(10):2080-96. doi: 10.3390/v4102080. Viruses. 2012. PMID: 23202454 Free PMC article. Review.
Cited by
- Genetic and metabolic signatures of Salmonella enterica subsp. enterica associated with animal sources at the pangenomic scale.
Vila Nova M, Durimel K, La K, Felten A, Bessières P, Mistou MY, Mariadassou M, Radomski N. Vila Nova M, et al. BMC Genomics. 2019 Nov 6;20(1):814. doi: 10.1186/s12864-019-6188-x. BMC Genomics. 2019. PMID: 31694533 Free PMC article. - Malaria protection due to sickle haemoglobin depends on parasite genotype.
Band G, Leffler EM, Jallow M, Sisay-Joof F, Ndila CM, Macharia AW, Hubbart C, Jeffreys AE, Rowlands K, Nguyen T, Gonçalves S, Ariani CV, Stalker J, Pearson RD, Amato R, Drury E, Sirugo G, d'Alessandro U, Bojang KA, Marsh K, Peshu N, Saelens JW, Diakité M, Taylor SM, Conway DJ, Williams TN, Rockett KA, Kwiatkowski DP. Band G, et al. Nature. 2022 Feb;602(7895):106-111. doi: 10.1038/s41586-021-04288-3. Epub 2021 Dec 9. Nature. 2022. PMID: 34883497 Free PMC article. - Lessons Learnt From Using the Machine Learning Random Forest Algorithm to Predict Virulence in Streptococcus pyogenes.
Buckley SJ, Harvey RJ. Buckley SJ, et al. Front Cell Infect Microbiol. 2021 Dec 24;11:809560. doi: 10.3389/fcimb.2021.809560. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 35004362 Free PMC article. Review. - Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives.
Abel L, Fellay J, Haas DW, Schurr E, Srikrishna G, Urbanowski M, Chaturvedi N, Srinivasan S, Johnson DH, Bishai WR. Abel L, et al. Lancet Infect Dis. 2018 Mar;18(3):e64-e75. doi: 10.1016/S1473-3099(17)30623-0. Epub 2017 Oct 27. Lancet Infect Dis. 2018. PMID: 29111156 Free PMC article. Review. - Correcting for Population Stratification Reduces False Positive and False Negative Results in Joint Analyses of Host and Pathogen Genomes.
Naret O, Chaturvedi N, Bartha I, Hammer C, Fellay J; Swiss HIV Cohort Study (SHCS). Naret O, et al. Front Genet. 2018 Jul 30;9:266. doi: 10.3389/fgene.2018.00266. eCollection 2018. Front Genet. 2018. PMID: 30105048 Free PMC article.
References
- Almeida CA, Bronke C, Roberts SG, McKinnon E, Keane NM, Chopra A, Kadie C, et al. 2011. Translation of HLA-HIV associations to the cellular level: HIV adapts to inflate CD8 T cell responses against Nef and HLA-adapted variant epitopes. J Immunol 187:2502–13. doi: 10.4049/jimmunol.1100691. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous