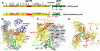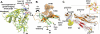Crystal structure of a soluble cleaved HIV-1 envelope trimer - PubMed (original) (raw)
. 2013 Dec 20;342(6165):1477-83.
doi: 10.1126/science.1245625. Epub 2013 Oct 31.
Albert Cupo, Devin Sok, Robyn L Stanfield, Dmitry Lyumkis, Marc C Deller, Per-Johan Klasse, Dennis R Burton, Rogier W Sanders, John P Moore, Andrew B Ward, Ian A Wilson
Affiliations
- PMID: 24179159
- PMCID: PMC3886632
- DOI: 10.1126/science.1245625
Crystal structure of a soluble cleaved HIV-1 envelope trimer
Jean-Philippe Julien et al. Science. 2013.
Abstract
HIV-1 entry into CD4(+) target cells is mediated by cleaved envelope glycoprotein (Env) trimers that have been challenging to characterize structurally. Here, we describe the crystal structure at 4.7 angstroms of a soluble, cleaved Env trimer that is stabilized and antigenically near-native (termed the BG505 SOSIP.664 gp140 trimer) in complex with a potent broadly neutralizing antibody, PGT122. The structure shows a prefusion state of gp41, the interaction between the component gp120 and gp41 subunits, and how a close association between the gp120 V1/V2/V3 loops stabilizes the trimer apex around the threefold axis. The complete epitope of PGT122 on the trimer involves gp120 V1, V3, and several surrounding glycans. This trimer structure advances our understanding of how Env functions and is presented to the immune system, and provides a blueprint for structure-based vaccine design.
Figures
Fig. 1. Overall architecture of a soluble, cleaved, recombinant HIV-1 Env trimer in complex with bnAb PGT 122
(A) Schematic of the soluble, cleaved, recombinant HIV-1 Env BG505 SOSIP.664 construct in comparison to full-length gp160. N-linked glycans are shown and numbered on their respective Asn residues. The constant (C1-C5) and variable (V1-V5) regions in gp120 and the fusion peptide (FP), HR1 and HR2 helices, membrane proximal external region (MPER), transmembrane (TM) and cytoplasmic (CT) elements in gp41 are indicated. The SOSIP mutations are shown in red, as well as the added N332 glycan site. The color coding is as in B. (B) Side view of the soluble Env trimer complex with PGT122 showing two of the three Env gp140 protomers associated with PGT122 Fab (blue). A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around the leftmost gp140 protomer. The membrane to which gp41 is attached would be at the bottom of the figure. (C) Side view of the Env trimer. For one of the three protomers on Env, core gp120 is shown in yellow, whereas V1/V2 and V3 regions are highlighted in orange and red, respectively. The main gp41 helical elements are colored in different shades of green. Protein components are rendered according to their secondary structure and glycans are depicted as spheres. (D) View of Env down the trimer axis. Loops of high variability in gp120 (V1-V5) all map to the periphery of the trimer and are labeled. Glycans have been omitted for clarity. Dashed lines indicate the location of gp120 V2 and V4 loops for which electron density was absent or ambiguous. The figure was generated with Pymol (63).
Fig. 2. Comparison of gp120 and components as observed in high-resolution crystal structures and in the soluble HIV-1 Env trimer
(A) High-resolution crystal structure of core gp120 (PDB ID: 3JWD, (5), pale green) is superimposed on the gp120 component of the soluble, cleaved SOSIP.664 trimer crystal structure (yellow). A longer α1 helix (gp120 residues 99-117) likely contributes to rearrangement in the bridging sheet, particularly in β2 and β3, which extend into V1/V2 atop the trimer. The gp120 β2-proximal residues 115-125 are highlighted in blue and brown in core gp120 and trimeric gp140, respectively. A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around the α1 helix. (B) Superimposition of the scaffolded gp120 V1/V2 crystal structure (PDB ID: 3U4E, pale green) on V1/V2 in the trimer crystal structure (orange). There are differences in V1 and in the β2 and β3 connecting strands. Electron density for carbohydrates at gp120 N156 and N160 is shown as a 2Fo-Fc gray mesh contoured at 1.0 σ. (C) Structural arrangement of gp120 V1/V2 (orange) and V3 (red) in the context of the trimer. A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around V1/V2 and V3 elements. All structures are depicted according to secondary structure elements, with glycans depicted as yellow spheres and the Cys126-Cys196 disulfide bond colored blue. The figure was generated with Pymol (63).
Fig. 3. Structural organization of gp41 in the soluble cleaved HIV-1 Env trimer
(A) Overall arrangement of gp41 elements from one protomer is shown in a gray 2Fo-Fc electron density map contoured at 1.0 σ. Carbohydrates are shown as spheres. Dashed lines delineate connecting electron density for which a chain trace and secondary structure determination was ambiguous at this moderate resolution. (B) Regions of contact between the gp120 inner domain and the gp41 central helix. The inset shows hydrophobic residues in the gp120 high-resolution crystal structure from C1, α0 and loop A that line the interface with gp41 HR1. (C) Surface rendering of two of the three HIV-1 Env and Influenza hemagglutinin (HA) (PDB ID: 4FNK) protomers (back protomer omitted) emphasizes the similarity in position and size of a small, central inter-protomer opening that presumably facilitates conformational changes during the fusion process. (D) Comparison between the structures of HIV-1 Env and Influenza HA (PDB ID: 4FNK), the prototype type I fusion protein. There are striking similarities in the position of structural elements in the two glycoproteins. (E) The trimeric arrangement of gp41 HR1 in the post-fusion conformation (PDB ID: 2X7R, (50), beige) superimposes closely with the central HR1 in the soluble HIV-1 Env trimer, which is similar to the retention of the three-helix bundle at the top of the long HA2 helix in Influenza HA. All structures are depicted according to secondary structure elements. The figure was generated with Pymol (63).
Fig. 4. Complete structural definition of the PGT122 epitope
(A) In addition to the N332 glycan (yellow), the PGT122 bnAb recognizes both protein and glycan elements near the base of gp120 V1 (orange) and V3 (red) to mediate broad and potent HIV-1 neutralization. Heavy and light chain CDRs are shown as dark and light blue tubes, respectively. Electron density for oligomannose glycans (spheres) surrounding the PGT122 epitope is shown as a 2Fo-Fc gray mesh contoured at 1.0 σ. (B) Superimposition of the PGT122 epitope with glycans from PGT121 (PDB ID: 4JY4 and 4FQC, (23, 55)) and PG16 (PDB ID: 4DQO, (6)) liganded crystal structures. PGT122 binding is compatible with the involvement of complex/hybrid glycans at gp120 N137 and N156/173 (green) when the trimer is expressed in human cells capable of making this type of glycan. The figure was generated with Pymol (63). (C) The use of glycan knock-out mutants of HIV-1 BG505 pseudoviruses reveals the importance of glycans at N137, N156 and N301 for PGT122 recognition and neutralization. These glycans are part of the PGT122 epitope in the trimer crystal structure to varying extents.
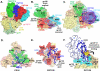
Fig. 5. Revisiting glycan-dependent epitopes of bnAbs in the context of the soluble Env trimer structure
(A) Superimposition of the co-crystal structures of PG16 (green), PGT128 (red) and PGT135 (cyan) on the PGT122 (blue)-Env trimer crystal structure reveals that these glycan-dependent epitopes overlap. (B) Co-crystal structures of glycan-dependent bnAbs in complex with monomeric gp120 or scaffolded gp120 elements provide crucial, but incomplete, details of their epitopes and, hence, how they recognize Env. The various bnAb epitopes are colored individually on the surfaces, with overlapping elements in purple. (C) Superimposition of the PG9/16, PGT128 and PGT135 co-crystal structures on the soluble trimer-PGT122 co-crystal structure reveals that these glycan-dependent bnAbs have expanded epitopes, which creates overlapping sites of vulnerability on HIV-1 Env. (D) Model of the expanded PG16 epitope on Env. Antibody interactions additional to those previously described in the PG9/PG16-V1/V2-scaffold co-crystal structures (4, 6) are predicted to occur with gp120 V3, as well as with residues N160 and N197 of the adjacent protomer. Protomers are denoted with P1, P2, P3 subscripts. (E). Model of the expanded PGT128 epitope includes additional interactions with glycans at N137 and N156 mediated via the PGT128 light chain (light blue). (F) PGT135 would clash (yellow star) with the gp120 V1 conformation recognized by PGT122, based on superimposition of the PGT135-core gp120 co-crystal structure (1). A slight re-orientation of gp120 V1 (green) would allow PGT135 to interact, consistent with the hypothesis that the V1 loop is flexible enough to permit different modes of bnAb interaction. Heavy and light chains are shown as dark and light blue tubes, respectively. The figure was generated with Pymol (63).
Similar articles
- Conformational States of a Soluble, Uncleaved HIV-1 Envelope Trimer.
Liu Y, Pan J, Cai Y, Grigorieff N, Harrison SC, Chen B. Liu Y, et al. J Virol. 2017 Apr 28;91(10):e00175-17. doi: 10.1128/JVI.00175-17. Print 2017 May 15. J Virol. 2017. PMID: 28250125 Free PMC article. - Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein.
Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. Klasse PJ, et al. J Virol. 2013 Sep;87(17):9873-85. doi: 10.1128/JVI.01226-13. Epub 2013 Jul 3. J Virol. 2013. PMID: 23824824 Free PMC article. - Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.
Kong L, Torrents de la Peña A, Deller MC, Garces F, Sliepen K, Hua Y, Stanfield RL, Sanders RW, Wilson IA. Kong L, et al. Acta Crystallogr D Biol Crystallogr. 2015 Oct;71(Pt 10):2099-108. doi: 10.1107/S1399004715013917. Epub 2015 Sep 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 26457433 Free PMC article. - HIV-1 envelope glycoprotein structure.
Merk A, Subramaniam S. Merk A, et al. Curr Opin Struct Biol. 2013 Apr;23(2):268-76. doi: 10.1016/j.sbi.2013.03.007. Epub 2013 Apr 18. Curr Opin Struct Biol. 2013. PMID: 23602427 Free PMC article. Review. - Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.
Liu Q, Zhang P, Lusso P. Liu Q, et al. Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
Cited by
- Stoichiometry of HIV-1 Envelope Glycoprotein Protomers with Changes That Stabilize or Destabilize the Pretriggered Conformation.
Zhang Z, Anang S, Wang Q, Nguyen HT, Chen HC, Chiu TJ, Yang D, Smith AB 3rd, Sodroski JG. Zhang Z, et al. bioRxiv [Preprint]. 2024 Oct 25:2024.10.25.620268. doi: 10.1101/2024.10.25.620268. bioRxiv. 2024. PMID: 39484577 Free PMC article. Preprint. - Computational design of prefusion-stabilized Herpesvirus gB trimers.
McCallum M, Veesler D. McCallum M, et al. bioRxiv [Preprint]. 2024 Oct 24:2024.10.23.619923. doi: 10.1101/2024.10.23.619923. bioRxiv. 2024. PMID: 39484573 Free PMC article. Preprint. - Potent broadly neutralizing antibodies mediate efficient antibody-dependent phagocytosis of HIV-infected cells.
Snow BJ, Keles NK, Grunst MW, Janaka SK, Behrens RT, Evans DT. Snow BJ, et al. PLoS Pathog. 2024 Oct 28;20(10):e1012665. doi: 10.1371/journal.ppat.1012665. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39466835 Free PMC article. - Antiretroviral Therapy with Ritonavir-Boosted Atazanavir- and Lopinavir-Containing Regimens Correlates with Diminished HIV-1 Neutralization.
Yuste E, Gil H, Garcia F, Sanchez-Merino V. Yuste E, et al. Vaccines (Basel). 2024 Oct 17;12(10):1176. doi: 10.3390/vaccines12101176. Vaccines (Basel). 2024. PMID: 39460342 Free PMC article. - Cryo-electron microscopy in the study of virus entry and infection.
Dutta M, Acharya P. Dutta M, et al. Front Mol Biosci. 2024 Jul 24;11:1429180. doi: 10.3389/fmolb.2024.1429180. eCollection 2024. Front Mol Biosci. 2024. PMID: 39114367 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- R01 AI33292/AI/NIAID NIH HHS/United States
- P01 AI82362/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- P41 GM103310/GM/NIGMS NIH HHS/United States
- U54 GM094586/GM/NIGMS NIH HHS/United States
- P41 RR001209/RR/NCRR NIH HHS/United States
- R37 AI036082/AI/NIAID NIH HHS/United States
- R37 AI36082/AI/NIAID NIH HHS/United States
- P01 AI082362/AI/NIAID NIH HHS/United States
- P41RR001209/RR/NCRR NIH HHS/United States
- R01 AI084817/AI/NIAID NIH HHS/United States
- CAPMC/ CIHR/Canada
- GM103310/GM/NIGMS NIH HHS/United States
- R01 AI033292/AI/NIAID NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous