Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs - PubMed (original) (raw)
. 2013 Nov 7;93(5):865-75.
doi: 10.1016/j.ajhg.2013.10.005. Epub 2013 Oct 31.
Jian Yang, Anna Vinkhuyzen, Joseph E Powell, Gonneke Willemsen, Jouke-Jan Hottenga, Abdel Abdellaoui, Massimo Mangino, Ana M Valdes, Sarah E Medland, Pamela A Madden, Andrew C Heath, Anjali K Henders, Dale R Nyholt, Eco J C de Geus, Patrik K E Magnusson, Erik Ingelsson, Grant W Montgomery, Timothy D Spector, Dorret I Boomsma, Nancy L Pedersen, Nicholas G Martin, Peter M Visscher
Affiliations
- PMID: 24183453
- PMCID: PMC3965855
- DOI: 10.1016/j.ajhg.2013.10.005
Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs
Gibran Hemani et al. Am J Hum Genet. 2013.
Abstract
Evidence that complex traits are highly polygenic has been presented by population-based genome-wide association studies (GWASs) through the identification of many significant variants, as well as by family-based de novo sequencing studies indicating that several traits have a large mutational target size. Here, using a third study design, we show results consistent with extreme polygenicity for body mass index (BMI) and height. On a sample of 20,240 siblings (from 9,570 nuclear families), we used a within-family method to obtain narrow-sense heritability estimates of 0.42 (SE = 0.17, p = 0.01) and 0.69 (SE = 0.14, p = 6 × 10(-)(7)) for BMI and height, respectively, after adjusting for covariates. The genomic inflation factors from locus-specific linkage analysis were 1.69 (SE = 0.21, p = 0.04) for BMI and 2.18 (SE = 0.21, p = 2 × 10(-10)) for height. This inflation is free of confounding and congruent with polygenicity, consistent with observations of ever-increasing genomic-inflation factors from GWASs with large sample sizes, implying that those signals are due to true genetic signals across the genome rather than population stratification. We also demonstrate that the distribution of the observed test statistics is consistent with both rare and common variants underlying a polygenic architecture and that previous reports of linkage signals in complex traits are probably a consequence of polygenic architecture rather than the segregation of variants with large effects. The convergent empirical evidence from GWASs, de novo studies, and within-family segregation implies that family-based sequencing studies for complex traits require very large sample sizes because the effects of causal variants are small on average.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
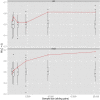
Figure 1
Q-Q Plots of Linkage Analyses For each cohort (left five panels), as well as the combined data set and the p values from the meta-analysis (the two right-most panels), Q-Q plots were produced for both BMI (top) and height (bottom) to demonstrate overall genomic inflation as being a departure from the expectation (x = y line). 95% confidence intervals are shaded in gray, and GC values for each cell represent genomic inflation. Markers are pruned to be 20 cM apart for reducing correlations between tests (for Q-Q plots on all markers, see Figure S12). From left to right, cohorts are TwinsUK, Netherlands Twin Registry, TWINGENE, Framingham, and QIMR. The sixth panel (n = 20,240) shows the results from the combined analysis, and the seventh panel represents the meta-analysis of all five cohorts.
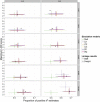
Figure 2
Proportion of Markers with Positive Heritability Estimates P(hm2>0) For each cohort (five bars to the left) and for the combined sample set (bar furthest to the right), an independent permutation analysis was performed such that the entire family of tests was rerun with QISP phenotypes randomly relabeled from QISP genomic IBD scores. Thus, each box-and-whisker plot represents the distribution of P(hm2>0) values from the 100 permutations per cohort; red points represent the P(hm2>0) achieved in the true linkage scans.
Figure 3
Simulations of Genetic Effect Models Expected results for various polygenic models are compared to observed results from true linkage analyses. Maximum LOD scores (y axis) from linkage scans are plotted against P(hm2>0) (x axis). Rows of panels correspond to sample sizes, representing different cohort sizes (top five rows) and the combined data set (bottom row). Columns of panels correspond to simulated _h_2; the left column represents BMI, and the right column represents height. Each model × sample size × heritability combination was replicated 100 times, and error bars represent 95% confidence intervals. Genetic models are as follows: Null, no genetic effects; C1, oligogenic where a single common QTL exists per chromosome; C2, polygenic with common SNPs uniformly distributed throughout the genome; C3, polygenic with common SNPs clustering across the genome; R1, polygenic with rare SNPs uniformly distributed throughout the genome; and R2, polygenic with rare SNPs clustering across the genome. It is shown that different polygenic architectures have nearly identical properties in linkage analysis and that they are consistent with the results for BMI and height.
Figure 4
Linkage Analysis for BMI and Height LOD scores for BMI across all autosomes are shown on the top row, and height is shown on the bottom row. The traditional threshold of LOD = 3.3 is depicted by solid lines, and the more stringent empirical threshold based on simulations of polygenic models is shown by dotted lines (α = 0.05, see Figure S16). Blue points represent GWAS hits from the GIANT study (different shades of blue correspond to different log10 p values). There is no correlation between linkage signals and GWAS signals (Figure S18).
Similar articles
- A population genetic interpretation of GWAS findings for human quantitative traits.
Simons YB, Bullaughey K, Hudson RR, Sella G. Simons YB, et al. PLoS Biol. 2018 Mar 16;16(3):e2002985. doi: 10.1371/journal.pbio.2002985. eCollection 2018 Mar. PLoS Biol. 2018. PMID: 29547617 Free PMC article. - Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals.
Chan Y, Holmen OL, Dauber A, Vatten L, Havulinna AS, Skorpen F, Kvaløy K, Silander K, Nguyen TT, Willer C, Boehnke M, Perola M, Palotie A, Salomaa V, Hveem K, Frayling TM, Hirschhorn JN, Weedon MN. Chan Y, et al. PLoS Genet. 2011 Dec;7(12):e1002439. doi: 10.1371/journal.pgen.1002439. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242009 Free PMC article. - A Selection Operator for Summary Association Statistics Reveals Allelic Heterogeneity of Complex Traits.
Ning Z, Lee Y, Joshi PK, Wilson JF, Pawitan Y, Shen X. Ning Z, et al. Am J Hum Genet. 2017 Dec 7;101(6):903-912. doi: 10.1016/j.ajhg.2017.09.027. Am J Hum Genet. 2017. PMID: 29198721 Free PMC article. - The omnigenic model and polygenic prediction of complex traits.
Mathieson I. Mathieson I. Am J Hum Genet. 2021 Sep 2;108(9):1558-1563. doi: 10.1016/j.ajhg.2021.07.003. Epub 2021 Jul 30. Am J Hum Genet. 2021. PMID: 34331855 Free PMC article. Review. - Estimation and partition of heritability in human populations using whole-genome analysis methods.
Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM. Vinkhuyzen AA, et al. Annu Rev Genet. 2013;47:75-95. doi: 10.1146/annurev-genet-111212-133258. Epub 2013 Aug 22. Annu Rev Genet. 2013. PMID: 23988118 Free PMC article. Review.
Cited by
- Genetic associations with micronutrient levels identified in immune and gastrointestinal networks.
Morine MJ, Monteiro JP, Wise C, Teitel C, Pence L, Williams A, Ning B, McCabe-Sellers B, Champagne C, Turner J, Shelby B, Bogle M, Beger RD, Priami C, Kaput J. Morine MJ, et al. Genes Nutr. 2014 Jul;9(4):408. doi: 10.1007/s12263-014-0408-4. Epub 2014 May 31. Genes Nutr. 2014. PMID: 24879315 Free PMC article. - Gene-obesogenic environment interactions in the UK Biobank study.
Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G, Joost S, Guessous I, Murray A, Strachan DP, Kutalik Z, Weedon MN, Frayling TM. Tyrrell J, et al. Int J Epidemiol. 2017 Apr 1;46(2):559-575. doi: 10.1093/ije/dyw337. Int J Epidemiol. 2017. PMID: 28073954 Free PMC article. - Novel epigenetic determinants of type 2 diabetes in Mexican-American families.
Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW Jr, Göring HH, Cole SA, Comuzzie AG, Almasy L, Mahaney MC, Curran JE, Blangero J, Carless MA. Kulkarni H, et al. Hum Mol Genet. 2015 Sep 15;24(18):5330-44. doi: 10.1093/hmg/ddv232. Epub 2015 Jun 22. Hum Mol Genet. 2015. PMID: 26101197 Free PMC article. - Reduced signal for polygenic adaptation of height in UK Biobank.
Berg JJ, Harpak A, Sinnott-Armstrong N, Joergensen AM, Mostafavi H, Field Y, Boyle EA, Zhang X, Racimo F, Pritchard JK, Coop G. Berg JJ, et al. Elife. 2019 Mar 21;8:e39725. doi: 10.7554/eLife.39725. Elife. 2019. PMID: 30895923 Free PMC article. - Analysis of the genetic basis of height in large Jewish nuclear families.
Zeevi D, Bloom JS, Sadhu MJ, Ben Yehuda A, Zangen D, Levy-Lahad E, Kruglyak L. Zeevi D, et al. PLoS Genet. 2019 Jul 8;15(7):e1008082. doi: 10.1371/journal.pgen.1008082. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31283753 Free PMC article.
References
- Swinburn B.A., Sacks G., Hall K.D., McPherson K., Finegood D.T., Moodie M.L., Gortmaker S.L. The global obesity pandemic: shaped by global drivers and local environments. Lancet. 2011;378:804–814. - PubMed
- Schvey N.A., Puhl R.M., Levandoski K.A., Brownell K.D. The influence of a defendant’s body weight on perceptions of guilt. Int. J. Obes. (Lond.) 2013;37:1275–1281. - PubMed
- Puhl R.M., Peterson J.L., Luedicke J. Weight-based victimization: bullying experiences of weight loss treatment-seeking youth. Pediatrics. 2013;131:e1–e9. - PubMed
Publication types
MeSH terms
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- AA13320/AA/NIAAA NIH HHS/United States
- GM099568/GM/NIGMS NIH HHS/United States
- DA12854/DA/NIDA NIH HHS/United States
- 1RC2 MH089995‐01/MH/NIMH NIH HHS/United States
- R01D0042157‐01A/PHS HHS/United States
- U24 MH068457‐06/MH/NIMH NIH HHS/United States
- N01‐HC‐25195/HC/NHLBI NIH HHS/United States
- N02‐HL‐64278/HL/NHLBI NIH HHS/United States
- GM075091/GM/NIGMS NIH HHS/United States
- AA07535/AA/NIAAA NIH HHS/United States
- AA13326/AA/NIAAA NIH HHS/United States
- AA13321/AA/NIAAA NIH HHS/United States
- AA10248/AA/NIAAA NIH HHS/United States
- 1RC2MH089951‐01/MH/NIMH NIH HHS/United States
- DK U01‐066134/DK/NIDDK NIH HHS/United States
- AA014041/AA/NIAAA NIH HHS/United States
- R01 GM075091/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources